c:ms:2025:lecture_note_week_04
This is an old revision of the document!
Table of Contents
Variance
Why n-1 for 모집단
# variance # why we use n-1 when calculating # population variance (instead of N) a <- rnorm2(100000000, 100, 10) a.mean <- mean(a) ss <- sum((a-a.mean)^2) n <- length(a) df <- n-1 ss/n ss/df
Variance output
> # variance > # why we use n-1 when calculating > # population variance (instead of N) > a <- rnorm2(100000000, 100, 10) > a.mean <- mean(a) > ss <- sum((a-a.mean)^2) > n <- length(a) > df <- n-1 > ss/n [1] 100 > ss/df [1] 100 >
rule of SD
# standard deviation 68, 85, 99 rule one.sd <- .68 two.sd <- .95 thr.sd <- .99 1-one.sd (1-one.sd)/2 qnorm(0.16) qnorm(1-0.16) 1-two.sd (1-two.sd)/2 qnorm(0.025) qnorm(0.975) 1-thr.sd (1-thr.sd)/2 qnorm(0.005) qnorm(1-0.005)
rule of SD output
> # standard deviation 68, 85, 99 rule > one.sd <- .68 > two.sd <- .95 > thr.sd <- .99 > 1-one.sd [1] 0.32 > (1-one.sd)/2 [1] 0.16 > qnorm(0.16) [1] -0.9944579 > qnorm(1-0.16) [1] 0.9944579 > > 1-two.sd [1] 0.05 > (1-two.sd)/2 [1] 0.025 > qnorm(0.025) [1] -1.959964 > qnorm(0.975) [1] 1.959964 > > 1-thr.sd [1] 0.01 > (1-thr.sd)/2 [1] 0.005 > qnorm(0.005) [1] -2.575829 > qnorm(1-0.005) [1] 2.575829 > >
Sampling distribution
OR the distribution of sample means
# sampling distribution
n.ajstu <- 100000
mean.ajstu <- 100
sd.ajstu <- 10
set.seed(1024)
ajstu <- rnorm2(n.ajstu, mean=mean.ajstu, sd=sd.ajstu)
mean(ajstu)
sd(ajstu)
var(ajstu)
min.value <- min(ajstu)
max.value <- max(ajstu)
min.value
max.value
iter <- 10000 # # of sampling
n.4 <- 4
means4 <- rep (NA, iter)
for(i in 1:iter){
means4[i] = mean(sample(ajstu, n.4))
}
n.25 <- 25
means25 <- rep (NA, iter)
for(i in 1:iter){
means25[i] = mean(sample(ajstu, n.25))
}
n.100 <- 100
means100 <- rep (NA, iter)
for(i in 1:iter){
means100[i] = mean(sample(ajstu, n.100))
}
n.400 <- 400
means400 <- rep (NA, iter)
for(i in 1:iter){
means400[i] = mean(sample(ajstu, n.400))
}
n.900 <- 900
means900 <- rep (NA, iter)
for(i in 1:iter){
means900[i] = mean(sample(ajstu, n.900))
}
n.1600 <- 1600
means1600 <- rep (NA, iter)
for(i in 1:iter){
means1600[i] = mean(sample(ajstu, n.1600))
}
n.2500 <- 2500
means2500 <- rep (NA, iter)
for(i in 1:iter){
means2500[i] = mean(sample(ajstu, n.2500))
}
h4 <- hist(means4)
h25 <- hist(means25)
h100 <- hist(means100)
h400 <- hist(means400)
h900 <- hist(means900)
h1600 <- hist(means1600)
h2500 <- hist(means2500)
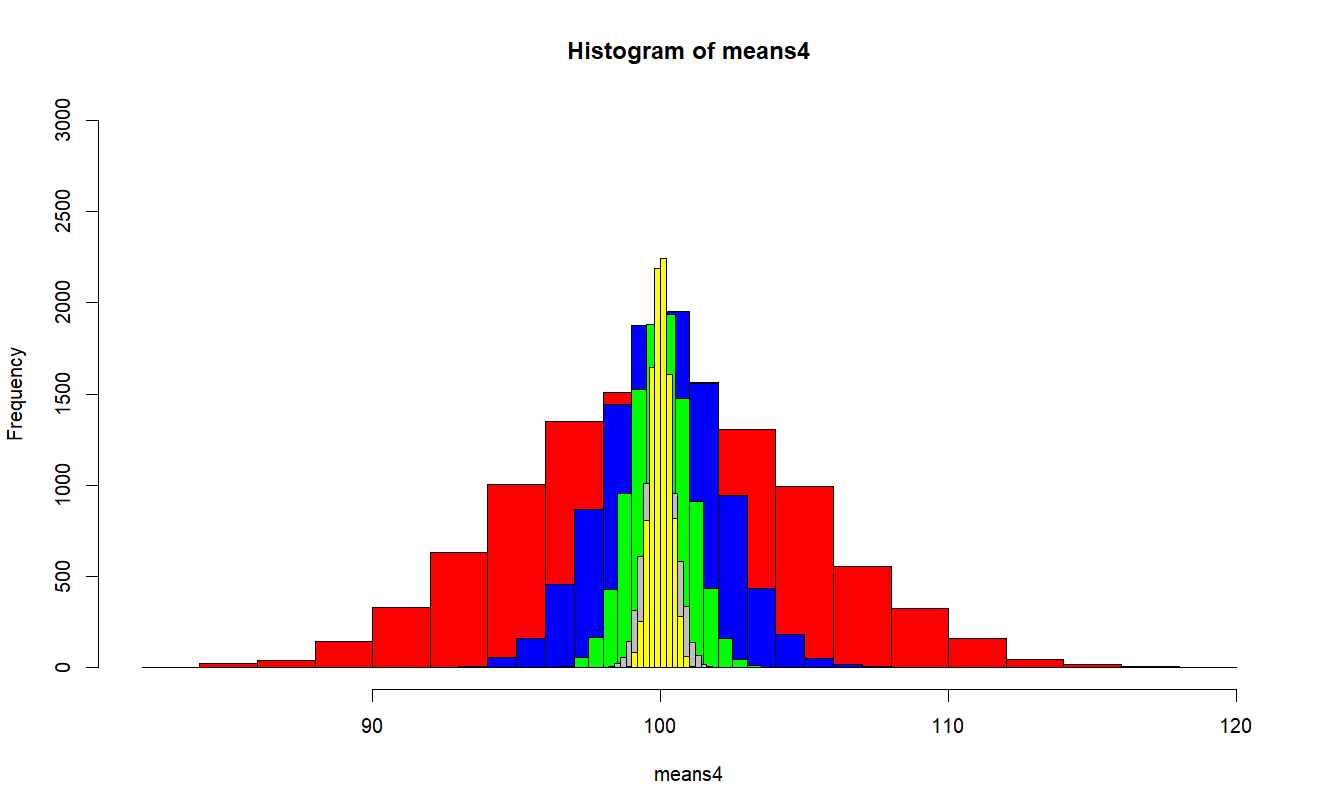
plot(h4, ylim=c(0,3000), col="red")
plot(h25, add = T, col="blue")
plot(h100, add = T, col="green")
plot(h400, add = T, col="grey")
plot(h900, add = T, col="yellow")
sss <- c(4,25,100,400,900,1600,2500) # sss sample sizes
ses <- rep (NA, length(sss)) # std errors
for(i in 1:length(sss)){
ses[i] = sqrt(var(ajstu)/sss[i])
}
ses.means4 <- sqrt(var(means4))
ses.means25 <- sqrt(var(means25))
ses.means100 <- sqrt(var(means100))
ses.means400 <- sqrt(var(means400))
ses.means900 <- sqrt(var(means900))
ses.means1600 <- sqrt(var(means1600))
ses.means2500 <- sqrt(var(means2500))
ses.real <- c(ses.means4, ses.means25,
ses.means100, ses.means400,
ses.means900, ses.means1600,
ses.means2500)
ses.real
ses
se.1 <- ses
se.2 <- 2 * ses
lower.s2 <- mean(ajstu)-se.2
upper.s2 <- mean(ajstu)+se.2
data.frame(cbind(sss, ses, ses.real, lower.s2, upper.s2))
min.value <- min(ajstu)
max.value <- max(ajstu)
min.value
max.value
# means25 분포에서 population의 가장 최소값인
# min.value가 나올 확률은 얼마나 될까?
m.25 <- mean(means25)
sd.25 <- sd(means25)
m.25
sd.25
pnorm(min.value, m.25, sd.25)
# 위처럼 최소값, 최대값의 probability를 가지고
# 확률을 구하는 것은 의미가 없음. 즉, 어떤 샘플의
# 평균이 원래 population에서 나왔는지를 따지는 것을
# 그 pop의 최소값으로 판단하는 것은 의미가 없음.
# 그 보다는 sd.25와 같은 standard deviation을 가지
# 고 보는 것이 타당함. 이 standard deviation 을
# standard error라고 부름
se.25 <- sd.25
se.25.2 <- 2 * sd.25
mean(means25)+c(-se.25.2, se.25.2)
# 위처럼 95% certainty를 가지고 구간을 판단하거나
# 아래 처럼 점수를 가지고 probability를 볼 수도
# 있음
pnorm(95.5, m.25, sd.25)
# 혹은
pnorm(95.5, m.25, sd.25) * 2
# 매우중요. 위의 이야기는 내가 25명의 샘플을 취
# 했을 때, 그 점수가 95.5가 나올 확률을 구해 본것
# 다른 예로
pnorm(106, m.25, sd.25, lower.tail = F)
# 혹은
2 * pnorm(106, m.25, sd.25, lower.tail = F)
Sampling distribution output
> # sampling distribution
> n.ajstu <- 100000
> mean.ajstu <- 100
> sd.ajstu <- 10
>
> set.seed(1024)
> ajstu <- rnorm2(n.ajstu, mean=mean.ajstu, sd=sd.ajstu)
>
> mean(ajstu)
[1] 100
> sd(ajstu)
[1] 10
> var(ajstu)
[,1]
[1,] 100
> min.value <- min(ajstu)
> max.value <- max(ajstu)
> min.value
[1] 57.10319
> max.value
[1] 141.9732
>
> iter <- 10000 # # of sampling
>
> n.4 <- 4
> means4 <- rep (NA, iter)
> for(i in 1:iter){
+ means4[i] = mean(sample(ajstu, n.4))
+ }
>
> n.25 <- 25
> means25 <- rep (NA, iter)
> for(i in 1:iter){
+ means25[i] = mean(sample(ajstu, n.25))
+ }
>
> n.100 <- 100
> means100 <- rep (NA, iter)
> for(i in 1:iter){
+ means100[i] = mean(sample(ajstu, n.100))
+ }
>
> n.400 <- 400
> means400 <- rep (NA, iter)
> for(i in 1:iter){
+ means400[i] = mean(sample(ajstu, n.400))
+ }
>
> n.900 <- 900
> means900 <- rep (NA, iter)
> for(i in 1:iter){
+ means900[i] = mean(sample(ajstu, n.900))
+ }
>
> n.1600 <- 1600
> means1600 <- rep (NA, iter)
> for(i in 1:iter){
+ means1600[i] = mean(sample(ajstu, n.1600))
+ }
>
> n.2500 <- 2500
> means2500 <- rep (NA, iter)
> for(i in 1:iter){
+ means2500[i] = mean(sample(ajstu, n.2500))
+ }
>
> h4 <- hist(means4)
> h25 <- hist(means25)
> h100 <- hist(means100)
> h400 <- hist(means400)
> h900 <- hist(means900)
> h1600 <- hist(means1600)
> h2500 <- hist(means2500)
>
>
> plot(h4, ylim=c(0,3000), col="red")
> plot(h25, add = T, col="blue")
> plot(h100, add = T, col="green")
> plot(h400, add = T, col="grey")
> plot(h900, add = T, col="yellow")
>
>
> sss <- c(4,25,100,400,900,1600,2500) # sss sample sizes
> ses <- rep (NA, length(sss)) # std errors
> for(i in 1:length(sss)){
+ ses[i] = sqrt(var(ajstu)/sss[i])
+ }
> ses.means4 <- sqrt(var(means4))
> ses.means25 <- sqrt(var(means25))
> ses.means100 <- sqrt(var(means100))
> ses.means400 <- sqrt(var(means400))
> ses.means900 <- sqrt(var(means900))
> ses.means1600 <- sqrt(var(means1600))
> ses.means2500 <- sqrt(var(means2500))
> ses.real <- c(ses.means4, ses.means25,
+ ses.means100, ses.means400,
+ ses.means900, ses.means1600,
+ ses.means2500)
> ses.real
[1] 4.9719142 2.0155741 0.9999527 0.5034433 0.3324414
[6] 0.2466634 0.1965940
>
> ses
[1] 5.0000000 2.0000000 1.0000000 0.5000000 0.3333333
[6] 0.2500000 0.2000000
> se.1 <- ses
> se.2 <- 2 * ses
>
> lower.s2 <- mean(ajstu)-se.2
> upper.s2 <- mean(ajstu)+se.2
> data.frame(cbind(sss, ses, ses.real, lower.s2, upper.s2))
sss ses ses.real lower.s2 upper.s2
1 4 5.0000000 4.9719142 90.00000 110.0000
2 25 2.0000000 2.0155741 96.00000 104.0000
3 100 1.0000000 0.9999527 98.00000 102.0000
4 400 0.5000000 0.5034433 99.00000 101.0000
5 900 0.3333333 0.3324414 99.33333 100.6667
6 1600 0.2500000 0.2466634 99.50000 100.5000
7 2500 0.2000000 0.1965940 99.60000 100.4000
>
> min.value <- min(ajstu)
> max.value <- max(ajstu)
> min.value
[1] 57.10319
> max.value
[1] 141.9732
> # means25 분포에서 population의 가장 최소값인
> # min.value가 나올 확률은 얼마나 될까?
> m.25 <- mean(means25)
> sd.25 <- sd(means25)
> m.25
[1] 100.0394
> sd.25
[1] 2.015574
> pnorm(min.value, m.25, sd.25)
[1] 5.414963e-101
> # 위처럼 최소값, 최대값의 probability를 가지고
> # 확률을 구하는 것은 의미가 없음. 즉, 어떤 샘플의
> # 평균이 원래 population에서 나왔는지를 따지는 것을
> # 그 pop의 최소값으로 판단하는 것은 의미가 없음.
> # 그 보다는 sd.25와 같은 standard deviation을 가지
> # 고 보는 것이 타당함. 이 standard deviation 을
> # standard error라고 부름
> se.25 <- sd.25
> se.25.2 <- 2 * sd.25
> mean(means25)+c(-se.25.2, se.25.2)
[1] 96.00822 104.07051
> # 위처럼 95% certainty를 가지고 구간을 판단하거나
> # 아래 처럼 점수를 가지고 probability를 볼 수도
> # 있음
> pnorm(95.5, m.25, sd.25)
[1] 0.01215658
> # 혹은
> pnorm(95.5, m.25, sd.25) * 2
[1] 0.02431317
> # 매우중요. 위의 이야기는 내가 25명의 샘플을 취
> # 했을 때, 그 점수가 95.5가 나올 확률을 구해 본것
> # 다른 예로
> pnorm(106, m.25, sd.25, lower.tail = F)
[1] 0.001551781
> # 혹은
> 2 * pnorm(106, m.25, sd.25, lower.tail = F)
[1] 0.003103562
>
>
>
c/ms/2025/lecture_note_week_04.1742789016.txt.gz · Last modified: by hkimscil