note.w02
This is an old revision of the document!
Table of Contents
Sampling Distribution and z-test
rm(list=ls())
rnorm2 <- function(n,mean,sd){
mean+sd*scale(rnorm(n))
}
se <- function(sample) {
sd(sample)/sqrt(length(sample))
}
ss <- function(x) {
sum((x-mean(x))^2)
}
################################
N.p <- 1000000
m.p <- 100
sd.p <- 10
p1 <- rnorm2(N.p, m.p, sd.p)
mean(p1)
sd(p1)
p2 <- rnorm2(N.p, m.p+20, sd.p)
mean(p2)
sd(p2)
m.p1 <- mean(p1)
sd.p1 <- sd(p1)
var(p1)
hist(p1)
hist(p1, breaks=50, col = rgb(1, 1, 1, 0.5),
main = "histogram of p1 and p2",)
abline(v=mean(p1), col="black", lwd=3)
hist(p2, add=T, breaks=50, col=rgb(1,1,.5,.5))
abline(v=mean(p2), col="red", lwd=3)
hist(p1, breaks=50, col=rgb(0,.5,.5,.5))
abline(v=mean(p1),lwd=2)
abline(v=m.p1-sd.p1, lwd=2)
abline(v=mean(p1)+sd(p1), lwd=2)
abline(v=c(m.p1-2*sd.p1, m.p1+2*sd.p1), lwd=2, col='red')
abline(v=c(m.p1-3*sd.p1, m.p1+3*sd.p1), lwd=2, col='green')
# area bet black = 68%
# between red = 95%
# between green = 99%
pnorm(m.p1+sd.p1, m.p1, sd.p1)
pnorm(m.p1-sd.p1, m.p1, sd.p1)
pnorm(m.p1+sd.p1, m.p1, sd.p1) -
pnorm(m.p1-sd.p1, m.p1, sd.p1)
pnorm(m.p1+2*sd.p1, m.p1, sd.p1) -
pnorm(m.p1-2*sd.p1, m.p1, sd.p1)
pnorm(m.p1+3*sd.p1, m.p1, sd.p1) -
pnorm(m.p1-3*sd.p1, m.p1, sd.p1)
pnorm(121, 100, 10) - pnorm(85, 100, 10)
m.p1
sd.p1
(m.p1-m.p1)/sd.p1
((m.p1-sd.p1) - m.p1) / sd.p1
(120-100)/10
pnorm(1)-pnorm(-1)
pnorm(2)-pnorm(-2)
pnorm(3)-pnorm(-3)
1-pnorm(-2)*2
pnorm(2)-pnorm(-2)
pnorm(120, 100, 10)
pnorm(2)-pnorm(-2)
zscore <- (120-100)/10
pnorm(zscore)-pnorm(-zscore)
zscore
# reminder.
pnorm(-1)
pnorm(1, 0, 1, lower.tail = F)
pnorm(110, 100, 10, lower.tail = F)
zscore <- (110-100)/10
pnorm(zscore, lower.tail = F)
pnorm(118, 100, 10, lower.tail = F)
pnorm(18/10, lower.tail = F)
z.p1 <- (p1-mean(p1))/sd(p1)
mean(z.p1)
round(mean(z.p1),10)
sd(z.p1)
pnorm(1.8)-pnorm(-1.8)
hist(z.p1, breaks=50, col=rgb(1,0,0,0))
abline(v=c(m.p1, -1.8, 1.8), col='red')
1-(pnorm(1.8)-pnorm(-1.8))
pnorm(1)-pnorm(-1)
1-(pnorm(-1)*2)
pnorm(2)-pnorm(-2)
1-(pnorm(-2)*2)
1-(pnorm(-3)*2)
#
hist(p1, breaks=50, col=rgb(.9,.9,.9,.9))
abline(v=mean(p1),lwd=2)
abline(v=mean(p1)-sd(p1), lwd=2)
abline(v=mean(p1)+sd(p1), lwd=2)
abline(v=c(m.p1-2*sd.p1, m.p1+2*sd.p1), lwd=2, col='red')
abline(v=c(m.p1-3*sd.p1, m.p1+3*sd.p1), lwd=2, col='green')
# 68%
a <- qnorm(.32/2)
b <- qnorm(1-.32/2)
c(a, b)
c(-1, 1)
# note that
.32/2
pnorm(-1)
qnorm(.32/2)
qnorm(pnorm(-1))
# 95%
c <- qnorm(.05/2)
d <- qnorm(1-.05/2)
c(c, d)
c(-2,2)
# 99%
e <- qnorm(.01/2)
f <- qnorm(1-.01/2)
c(e,f)
c(-3,3)
pnorm(b)-pnorm(a)
c(a, b)
pnorm(d)-pnorm(c)
c(c, d)
pnorm(f)-pnorm(e)
c(e, f)
qnorm(.5)
qnorm(1)
################################
s.size <- 10
means.temp <- c()
s1 <- sample(p1, s.size, replace = T)
mean(s1)
means.temp <- append(means.temp, mean(s1))
means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T)))
means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T)))
means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T)))
means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T)))
means.temp
iter <- 1000000
# means <- c()
means <- rep(NA, iter)
for (i in 1:iter) {
# means <- append(means, mean(sample(p1, s.size, replace = T)))
means[i] <- mean(sample(p1, s.size, replace = T))
}
length(means)
mean(means)
sd(means)
se.s <- sd(means)
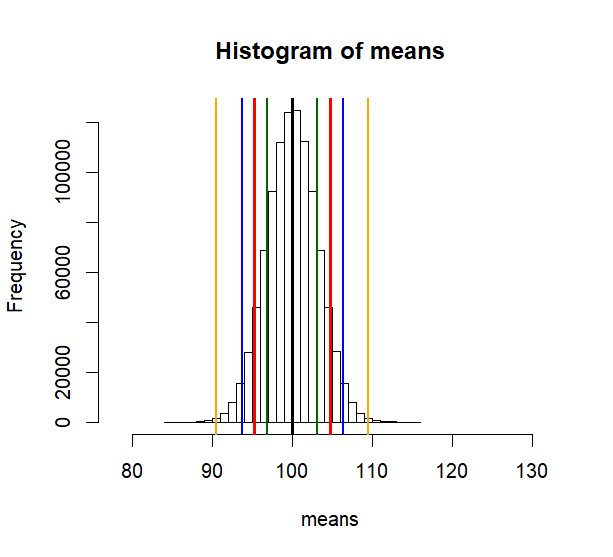
hist(means, breaks=50,
xlim = c(mean(means)-5*sd(means), mean(means)+10*sd(means)),
col=rgb(1, 1, 1, .5))
abline(v=mean(means), col="black", lwd=3)
# now we want to get sd of this distribution
lo1 <- mean(means)-se.s
hi1 <- mean(means)+se.s
lo2 <- mean(means)-2*se.s
hi2 <- mean(means)+2*se.s
lo3 <- mean(means)-3*se.s
hi3 <- mean(means)+3*se.s
abline(v=mean(means), col="black", lwd=2)
# abline(v=mean(p2), colo='darkgreen', lwd=2)
abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
col=c("red","red", "blue", "blue", "orange", "orange"),
lwd=2)
# meanwhile . . . .
se.s
se.z <- sqrt(var(p1)/s.size)
se.z
se.z <- c(se.z)
se.z
se.z.adjusted <- sqrt(var(s1)/s.size)
se.z.adjusted
# sd of sample means (sd(means))
# = se.s
# when iter value goes to
# infinite value:
# mean(means) = mean(p1)
# and
# sd(means) = sd(p1) / sqrt(s.size)
# that is, se.s = se.z
# This is called CLT (Central Limit Theorem)
# see http://commres.net/wiki/cetral_limit_theorem
mean(means)
mean(p1)
sd(means)
var(p1)
# remember we started talking sample size 10
sqrt(var(p1)/s.size)
se.z
sd(means)
se.s
se.z
# because CLT
loz1 <- mean(p1)-se.z
hiz1 <- mean(p1)+se.z
loz2 <- mean(p1)-2*se.z
hiz2 <- mean(p1)+2*se.z
loz3 <- mean(p1)-3*se.z
hiz3 <- mean(p1)+3*se.z
c(lo1, loz1)
c(lo2, loz2)
c(lo3, loz3)
c(hi1, hiz1)
c(hi2, hiz2)
c(hi3, hiz3)
hist(means, breaks=50,
xlim = c(mean(means)-5*sd(means), mean(means)+10*sd(means)),
col = rgb(1, 1, 1, .5))
abline(v=mean(means), col="black", lwd=3)
# abline(v=mean(p2), colo='darkgreen', lwd=3)
abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
col=c("darkgreen","darkgreen", "blue", "blue", "orange", "orange"),
lwd=2)
round(c(lo1, hi1))
round(c(lo2, hi2))
round(c(lo3, hi3))
round(c(loz1, hiz1))
round(c(loz2, hiz2))
round(c(loz3, hiz3))
m.sample.i.got <- mean(means)+ 1.5*sd(means)
m.sample.i.got
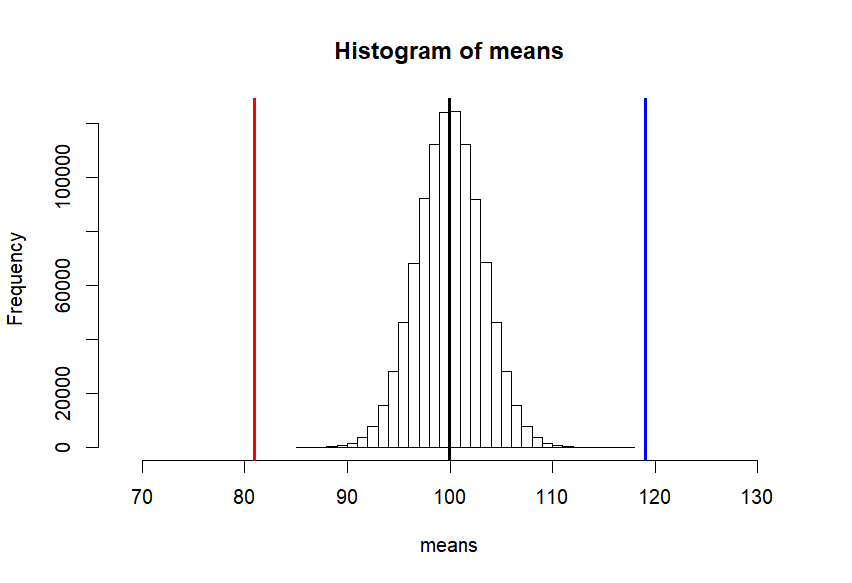
hist(means, breaks=30,
xlim = c(mean(means)-7*sd(means), mean(means)+10*sd(means)),
col = rgb(1, 1, 1, .5))
abline(v=mean(means), col="black", lwd=3)
abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
col=c("darkgreen","darkgreen", "blue", "blue", "orange", "orange"),
lwd=2)
abline(v=m.sample.i.got, col='red', lwd=3)
# what is the probablity of getting
# greater than
# m.sample.i.got?
m.sample.i.got
pnorm(m.sample.i.got, mean(means), sd(means), lower.tail = F)
pnorm(m.sample.i.got, mean(p1), se.z, lower.tail = F)
# then, what is the probabilty of getting
# greater than m.sample.i.got and
# less than corresponding value, which is
# mean(means) - m.sample.i.got - mean(means)
# (green line)
tmp <- mean(means) - (m.sample.i.got - mean(means))
abline(v=tmp, col='red', lwd=3)
2 * pnorm(m.sample.i.got, mean(p1), sd(means), lower.tail = F)
m.sample.i.got
### one more time
# this time, with a story
mean(p2)
sd(p2)
s.from.p2 <- sample(p2, s.size)
m.s.from.p2 <- mean(s.from.p2)
m.s.from.p2
se.s
se.z
sd(means)
m.k <- mean(s.from.p2)
se.k <- sd(s.from.p2)/sqrt(s.size)
tmp <- mean(means) - (m.s.from.p2
- mean(means))
tmp
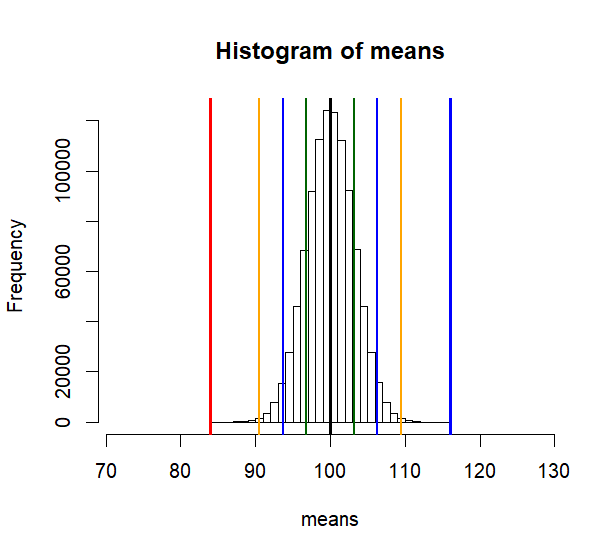
hist(means, breaks=30,
xlim = c(tmp-4*sd(means), m.s.from.p2+4*sd(means)),
col = rgb(1, 1, 1, .5))
abline(v=mean(means), col="black", lwd=3)
abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
col=c("darkgreen","darkgreen", "blue", "blue", "orange", "orange"),
lwd=2)
abline(v=m.s.from.p2, col='blue', lwd=3)
# what is the probablity of getting
# greater than
# m.sample.i.got?
m.s.from.p2
pnorm(m.s.from.p2, mean(p1), se.z, lower.tail = F)
pnorm(m.s.from.p2, m.k, se.k, lower.tail = F)
# then, what is the probabilty of getting
# greater than m.sample.i.got and
# less than corresponding value, which is
# mean(means) - m.sample.i.got - mean(means)
# (green line)
abline(v=tmp, col='red', lwd=3)
2 * pnorm(m.s.from.p2, mean(p1), se.z, lower.tail = F)
2 * pnorm(m.s.from.p2, m.k, se.k, lower.tail = F)
se.z
sd(s.from.p2)/sqrt(s.size)
se.z.adjusted <- sqrt(var(s.from.p2)/s.size)
se.z.adjusted
2 * pnorm(m.s.from.p2, mean(p1), se.z.adjusted,
lower.tail = F)
z.cal <- (m.s.from.p2 - mean(p1))/se.z.adjusted
z.cal
pnorm(z.cal, lower.tail = F)*2
pt(z.cal, 49, lower.tail = F)*2
t.test(s.from.p2, mu=mean(p1), var.equal = T)
output
1
> rm(list=ls())
> rnorm2 <- function(n,mean,sd){
+ mean+sd*scale(rnorm(n))
+ }
>
> se <- function(sample) {
+ sd(sample)/sqrt(length(sample))
+ }
>
> ss <- function(x) {
+ sum((x-mean(x))^2)
+ }
>
…………………………………………………………………
2
> ################################
> N.p <- 1000000
> m.p <- 100
> sd.p <- 10
>
> p1 <- rnorm2(N.p, m.p, sd.p)
> mean(p1)
[1] 100
> sd(p1)
[1] 10
>
> p2 <- rnorm2(N.p, m.p+5, sd.p)
> mean(p2)
[1] 105
> sd(p2)
[1] 10
>
> m.p1 <- mean(p1)
> sd.p1 <- sd(p1)
> var(p1)
[,1]
[1,] 100
>
>
> hist(p1, breaks=50, col = rgb(1, 1, 1, 0.5),
+ main = "histogram of p1 and p2",)
> abline(v=mean(p1), col="black", lwd=3)
> hist(p2, add=T, breaks=50, col=rgb(1,1,.5,.5))
> abline(v=mean(p2), col="red", lwd=3)
>
>
…………………………………………………………………
3
> hist(p1, breaks=50, col=rgb(0,.5,.5,.5)) > abline(v=mean(p1),lwd=2) > abline(v=mean(p1)-sd(p1), lwd=2) > abline(v=mean(p1)+sd(p1), lwd=2) > abline(v=c(m.p1-2*sd.p1, m.p1+2*sd.p1), lwd=2, col='red') > abline(v=c(m.p1-3*sd.p1, m.p1+3*sd.p1), lwd=2, col='green') > > # area bet black = 68% > # between red = 95% > # between green = 99% > > pnorm(m.p1+sd.p1, m.p1, sd.p1) [1] 0.8413447 > pnorm(m.p1-sd.p1, m.p1, sd.p1) [1] 0.1586553 > pnorm(m.p1+sd.p1, m.p1, sd.p1) - + pnorm(m.p1-sd.p1, m.p1, sd.p1) [1] 0.6826895 > > pnorm(m.p1+2*sd.p1, m.p1, sd.p1) - + pnorm(m.p1-2*sd.p1, m.p1, sd.p1) [1] 0.9544997 > > pnorm(m.p1+3*sd.p1, m.p1, sd.p1) - + pnorm(m.p1-3*sd.p1, m.p1, sd.p1) [1] 0.9973002 >
…………………………………………………………………
4
> m.p1 [1] 100 > sd.p1 [1] 10 > > (m.p1-m.p1)/sd.p1 [1] 0 > ((m.p1-sd.p1) - m.p1) / sd.p1 [1] -1 > (120-100)/10 [1] 2 > pnorm(1)-pnorm(-1) [1] 0.6826895 > pnorm(2)-pnorm(-2) [1] 0.9544997 > pnorm(3)-pnorm(3) [1] 0 > > 1-pnorm(-2)*2 [1] 0.9544997 > pnorm(2)-pnorm(-2) [1] 0.9544997 > > pnorm(120, 100, 10) [1] 0.9772499 > pnorm(2)-pnorm(-2) [1] 0.9544997 > > zscore <- (120-100)/10 > pnorm(zscore)-pnorm(-zscore) [1] 0.9544997 > zscore [1] 2 > > # reminder. > pnorm(-1) [1] 0.1586553 > pnorm(1, 0, 1, lower.tail = F) [1] 0.1586553 > pnorm(110, 100, 10, lower.tail = F) [1] 0.1586553 > zscore <- (110-100)/10 > pnorm(zscore, lower.tail = F) [1] 0.1586553 > > pnorm(118, 100, 10, lower.tail = F) [1] 0.03593032 > pnorm(18/10, lower.tail = F) [1] 0.03593032 >
…………………………………………………………………
5
> z.p1 <- (p1-mean(p1))/sd(p1) > mean(z.p1) [1] 8.189457e-18 > round(mean(z.p1),10) [1] 0 > sd(z.p1) [1] 1 > pnorm(1.8)-pnorm(-1.8) [1] 0.9281394 > > hist(z.p1, breaks=50, col=rgb(1,0,0,0)) > abline(v=c(m.p1, -1.8, 1.8), col='red') > 1-(pnorm(1.8)-pnorm(-1.8)) [1] 0.07186064 > > > pnorm(1)-pnorm(-1) [1] 0.6826895 > 1-(pnorm(-1)*2) [1] 0.6826895 > > pnorm(2)-pnorm(-2) [1] 0.9544997 > 1-(pnorm(-2)*2) [1] 0.9544997 > > 1-(pnorm(-3)*2) [1] 0.9973002 > > #
…………………………………………………………………
6
> hist(p1, breaks=50, col=rgb(.9,.9,.9,.9)) > abline(v=mean(p1),lwd=2) > abline(v=mean(p1)-sd(p1), lwd=2) > abline(v=mean(p1)+sd(p1), lwd=2) > abline(v=c(m.p1-2*sd.p1, m.p1+2*sd.p1), lwd=2, col='red') > abline(v=c(m.p1-3*sd.p1, m.p1+3*sd.p1), lwd=2, col='green') > > # 68% > a <- qnorm(.32/2) > b <- qnorm(1-.32/2) > c(a, b) [1] -0.9944579 0.9944579 > c(-1, 1) [1] -1 1 > # note that > .32/2 [1] 0.16 > pnorm(-1) [1] 0.1586553 > qnorm(.32/2) [1] -0.9944579 > qnorm(pnorm(-1)) [1] -1 > > # 95% > c <- qnorm(.05/2) > d <- qnorm(1-.05/2) > c(c, d) [1] -1.959964 1.959964 > c(-2,2) [1] -2 2 > > # 99% > e <- qnorm(.01/2) > f <- qnorm(1-.01/2) > c(e,f) [1] -2.575829 2.575829 > c(-3,3) [1] -3 3 > > > pnorm(b)-pnorm(a) [1] 0.68 > c(a, b) [1] -0.9944579 0.9944579 > pnorm(d)-pnorm(c) [1] 0.95 > c(c, d) [1] -1.959964 1.959964 > pnorm(f)-pnorm(e) [1] 0.99 > c(e, f) [1] -2.575829 2.575829 > > qnorm(.5) [1] 0 > qnorm(1) [1] Inf > >
…………………………………………………………………
7
> ################################ > s.size <- 50 > > means.temp <- c() > s1 <- sample(p1, s.size, replace = T) > mean(s1) [1] 98.76098 > means.temp <- append(means.temp, mean(s1)) > means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T))) > means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T))) > means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T))) > means.temp <- append(means.temp, mean(sample(p1, s.size, replace = T))) > means.temp [1] 98.76098 99.90935 99.29643 99.66014 101.93822 >
…………………………………………………………………
8
> iter <- 1000000
> # means <- c()
> means <- rep(NA, iter)
> for (i in 1:iter) {
+ # means <- append(means, mean(sample(p1, s.size, replace = T)))
+ means[i] <- mean(sample(p1, s.size, replace = T))
+ }
> length(means)
[1] 1000000
> mean(means)
[1] 99.99824
> sd(means)
[1] 1.414035
> se.s <- sd(means)
>
> hist(means, breaks=50,
+ xlim = c(mean(means)-5*sd(means), mean(means)+10*sd(means)),
+ col=rgb(1, 1, 1, .5))
> abline(v=mean(means), col="black", lwd=3)
> # now we want to get sd of this distribution
> lo1 <- mean(means)-se.s
> hi1 <- mean(means)+se.s
> lo2 <- mean(means)-2*se.s
> hi2 <- mean(means)+2*se.s
> lo3 <- mean(means)-3*se.s
> hi3 <- mean(means)+3*se.s
> abline(v=mean(means), col="black", lwd=2)
> # abline(v=mean(p2), colo='darkgreen', lwd=2)
> abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
+ col=c("red","red", "blue", "blue", "orange", "orange"),
+ lwd=2)
>
…………………………………………………………………
9
> # meanwhile . . . .
> se.s
[1] 1.414035
# se.s = sd(means)
# The below is from CLT
# see http://commres.org/wiki/central limit theorem
#
> se.z <- sqrt(var(p1)/s.size)
> se.z <- c(se.z)
> se.z
[1] 1.414214
>
> se.z.adjusted <- sqrt(var(s1)/s.size)
> se.z.adjusted
[1] 1.370464
>
> # sd of sample means (sd(means))
> # = se.s
>
> # when iter value goes to
> # infinite value:
> # mean(means) = mean(p1)
> # and
> # sd(means) = sd(p1) / sqrt(s.size)
> # that is, se.s = se.z
> # This is called CLT (Central Limit Theorem)
> # see http://commres.net/wiki/cetral_limit_theorem
>
> mean(means)
[1] 99.99824
> mean(p1)
[1] 100
> sd(means)
[1] 1.414035
> var(p1)
[,1]
[1,] 100
> # remember we started talking sample size 10
> sqrt(var(p1)/s.size)
[,1]
[1,] 1.414214
> se.z
[1] 1.414214
>
> sd(means)
[1] 1.414035
> se.s
[1] 1.414035
>
…………………………………………………………………
central limit theorem
$$\overline{X} \sim \displaystyle \text{N} \left(\mu, \dfrac{\sigma^{2}}{n} \right)$$
hypothesis testing
types of error
types of error
10
> # because of CLT we can use the > # below instead of > # mean(means)+-se.s > # > loz1 <- mean(p1)-se.z > hiz1 <- mean(p1)+se.z > loz2 <- mean(p1)-2*se.z > hiz2 <- mean(p1)+2*se.z > loz3 <- mean(p1)-3*se.z > hiz3 <- mean(p1)+3*se.z > > c(lo1, loz1) [1] 98.58421 98.58579 > c(lo2, loz2) [1] 97.17017 97.17157 > c(lo3, loz3) [1] 95.75614 95.75736 > > c(hi1, hiz1) [1] 101.4123 101.4142 > c(hi2, hiz2) [1] 102.8263 102.8284 > c(hi3, hiz3) [1] 104.2403 104.2426 > >
…………………………………………………………………
11
> hist(means, breaks=50,
+ xlim = c(mean(means)-5*sd(means), mean(means)+10*sd(means)),
+ col = rgb(1, 1, 1, .5))
> abline(v=mean(means), col="black", lwd=3)
> # abline(v=mean(p2), colo='darkgreen', lwd=3)
> abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
+ col=c("darkgreen","darkgreen", "blue", "blue", "orange", "orange"),
+ lwd=2)
>
> round(c(lo1, hi1))
[1] 99 101
> round(c(lo2, hi2))
[1] 97 103
> round(c(lo3, hi3))
[1] 96 104
>
> round(c(loz1, hiz1))
[1] 99 101
> round(c(loz2, hiz2))
[1] 97 103
> round(c(loz3, hiz3))
[1] 96 104
>
…………………………………………………………………
12
> m.sample.i.got <- mean(means)+ 1.5*sd(means) > m.sample.i.got [1] 102.1193 > > hist(means, breaks=30, + xlim = c(mean(means)-7*sd(means), mean(means)+10*sd(means)), + col = rgb(1, 1, 1, .5)) > abline(v=mean(means), col="black", lwd=3) > abline(v=m.sample.i.got, col='darkgreen', lwd=3) > > # what is the probablity of getting > # greater than > # m.sample.i.got? > m.sample.i.got [1] 102.1193 > pnorm(m.sample.i.got, mean(means), sd(means), lower.tail = F) [1] 0.0668072 > pnorm(m.sample.i.got, mean(p1), se.z, lower.tail = F) [1] 0.0669929 > > # then, what is the probabilty of getting > # greater than m.sample.i.got and > # less than corresponding value, which is > # mean(means) - m.sample.i.got - mean(means) > # (green line) > tmp <- mean(means) - (m.sample.i.got - mean(means)) > abline(v=tmp, col='red', lwd=3) > 2 * pnorm(m.sample.i.got, mean(p1), sd(means), lower.tail = F) [1] 0.1339368 > m.sample.i.got [1] 102.1193 >
13
> ### one more time
> # this time, with a story
> mean(p2)
[1] 120
> sd(p2)
[1] 10
> s.from.p2 <- sample(p2, s.size)
> m.s.from.p2 <- mean(s.from.p2)
> m.s.from.p2
[1] 116.0821
>
> se.s
[1] 3.166623
> se.z
[1] 3.162278
> sd(means)
[1] 3.166623
>
> m.k <- mean(s.from.p2)
> se.k <- sd(s.from.p2)/sqrt(s.size)
>
>
> tmp <- mean(means) - (m.s.from.p2
+ - mean(means))
> tmp
[1] 83.92356
>
> hist(means, breaks=30,
+ xlim = c(tmp-4*sd(means), m.s.from.p2+4*sd(means)),
+ col = rgb(1, 1, 1, .5))
> abline(v=mean(means), col="black", lwd=3)
> abline(v=c(lo1, hi1, lo2, hi2, lo3, hi3),
+ col=c("darkgreen","darkgreen", "blue", "blue", "orange", "orange"),
+ lwd=2)
> abline(v=m.s.from.p2, col='blue', lwd=3)
>
> # what is the probablity of getting
> # greater than
> # m.sample.i.got?
> m.s.from.p2
[1] 116.0821
> pnorm(m.s.from.p2, mean(p1), se.z, lower.tail = F)
[1] 1.832221e-07
> pnorm(m.s.from.p2, m.k, se.k, lower.tail = F)
[1] 0.5
> # then, what is the probabilty of getting
> # greater than m.sample.i.got and
> # less than corresponding value, which is
> # mean(means) - m.sample.i.got - mean(means)
> # (green line)
> abline(v=tmp, col='red', lwd=3)
> 2 * pnorm(m.s.from.p2, mean(p1), se.z, lower.tail = F)
[1] 3.664442e-07
>
> 2 * pnorm(m.s.from.p2, m.k, se.k, lower.tail = F)
[1] 1
>
>
> se.z
[1] 3.162278
> sd(s.from.p2)/sqrt(s.size)
[1] 3.209216
> se.z.adjusted <- sqrt(var(s.from.p2)/s.size)
> se.z.adjusted
[1] 3.209216
> 2 * pnorm(m.s.from.p2, mean(p1), se.z.adjusted,
+ lower.tail = F)
[1] 5.408382e-07
>
> z.cal <- (m.s.from.p2 - mean(p1))/se.z.adjusted
> z.cal
[1] 5.011228
> pnorm(z.cal, lower.tail = F)*2
[1] 5.408382e-07
>
>
> pt(z.cal, 49, lower.tail = F)*2
[1] 7.443756e-06
> t.test(s.from.p2, mu=mean(p1), var.equal = T)
One Sample t-test
data: s.from.p2
t = 5.0112, df = 9, p-value = 0.0007277
alternative hypothesis: true mean is not equal to 100
95 percent confidence interval:
108.8224 123.3419
sample estimates:
mean of x
116.0821
>
>
T-test sum up
rm(list=ls())
rnorm2 <- function(n,mean,sd){
mean+sd*scale(rnorm(n))
}
se <- function(sample) {
sd(sample)/sqrt(length(sample))
}
ss <- function(x) {
sum((x-mean(x))^2)
}
N.p <- 1000000
m.p <- 100
sd.p <- 10
p1 <- rnorm2(N.p, m.p, sd.p)
mean(p1)
sd(p1)
p2 <- rnorm2(N.p, m.p+10, sd.p)
mean(p2)
sd(p2)
hist(p1, breaks=50, col = rgb(1, 0, 0, 0.5),
main = "histogram of p1 and p2",)
abline(v=mean(p1), col="black", lwd=3)
hist(p2, breaks=50, add=TRUE, col = rgb(0, 0, 1, 0.5))
abline(v=mean(p2), col="red", lwd=3)
s.size <- 1000
s2 <- sample(p2, s.size)
mean(s2)
sd(s2)
se.z <- sqrt(var(p1)/s.size)
se.z <- c(se.z)
se.z.range <- c(-2*se.z,2*se.z)
se.z.range
mean(p1)+se.z.range
mean(s2)
z.cal <- (mean(s2) - mean(p1)) / se.z
z.cal
pnorm(z.cal, lower.tail = F) * 2
z.cal
# principles . . .
# distribution of sample means
iter <- 100000
means <- c()
for (i in 1:iter) {
m.of.s <- mean(sample(p1, s.size))
means <- append(means, m.of.s)
}
hist(means,
xlim = c(mean(means)-3*sd(means), mean(s2)+5),
col = rgb(1, 0, 0, 0.5))
abline(v=mean(p1), col="black", lwd=3)
abline(v=mean(s2),
col="blue", lwd=3)
lo1 <- mean(p1)-se.z
hi1 <- mean(p1)+se.z
lo2 <- mean(p1)-2*se.z
hi2 <- mean(p1)+2*se.z
abline(v=c(lo1, hi1, lo2, hi2),
col=c("green","green", "brown", "brown"),
lwd=2)
se.z
c(lo2, hi2)
pnorm(z.cal, lower.tail = F) * 2
# Note that we use sqrt(var(s2)/s.size)
# as our se value instread of
# sqrt(var(p1)/s.size)
# This is a common practice for R
# In fact, some z.test (made by someone)
# function uses the former rather than
# latter.
sqrt(var(p1)/s.size)
se.z
sqrt(var(s2)/s.size)
se(s2)
t.cal.os <- (mean(s2) - mean(p1))/ se(s2)
z.cal <- (mean(s2) - mean(p1))/ se.z
t.cal.os
z.cal
df.s2 <- length(s2)-1
df.s2
p.t.os <- pt(abs(t.cal.os), df.s2, lower.tail = F) * 2
p.t.os
t.out <- t.test(s2, mu=mean(p1))
library(BSDA)
z.out <- z.test(s2, p1, sigma.x = sd(s2), sigma.y = sd(p1))
z.out$statistic # se.z 대신에 se(s2) 값으로 구한 z 값
z.cal # se.z으로 (sqrt(var(p1)/s.size)값) 구한 z 값
t.out$statistic # se(s2)를 분모로 하여 구한 t 값
t.cal.os # se(s2)를 이용하여 손으로 구한 t 값
# But, after all, we use t.test method regardless of
# variation
hist(means,
xlim = c(mean(means)-3*sd(means), mean(s2)+5),
col = rgb(1, 0, 0, 0.5))
abline(v=mean(p1), col="black", lwd=3)
abline(v=mean(s2),
col="blue", lwd=3)
lo1 <- mean(p1)-se.z
hi1 <- mean(p1)+se.z
lo2 <- mean(p1)-2*se.z
hi2 <- mean(p1)+2*se.z
abline(v=c(lo1, hi1, lo2, hi2),
col=c("green","green", "brown", "brown"),
lwd=2)
# difference between black and blue line
# divided by
# se(s2) (= random difference)
# t.value
mean(s2)
mean(p1)
diff <- mean(s2)-mean(p1)
diff
se(s2)
diff/se(s2)
t.cal.os
########################
# 2 sample t-test
########################
# 가정. 아래에서 추출하는 두
# 샘플의 모집단의 파라미터를
# 모른다.
s1 <- sample(p1, s.size)
s2 <- sample(p2, s.size)
mean(s1)
mean(s2)
ss(s1)
ss(s2)
df.s1 <- length(s1)-1
df.s2 <- length(s2)-1
df.s1
df.s2
pooled.variance <- (ss(s1)+ss(s2))/(df.s1+df.s2)
pooled.variance
se.ts <- sqrt((pooled.variance/length(s1))+(pooled.variance/length(s2)))
se.ts
t.cal.ts <- (mean(s1)-mean(s2))/se.ts
t.cal.ts
p.val.ts <- pt(abs(t.cal.ts), df=df.s1+df.s2, lower.tail = F) * 2
p.val.ts
t.test(s1, s2, var.equal = T)
se(s1)
se(s2)
mean(s1)+c(-se(s1)*2, se(s1)*2)
mean(s2)+c(-se(s2)*2, se(s2)*2)
mean(p1)
mean(p2)
hist(s1, breaks=50,
col = rgb(1, 0, 0, 0.5))
hist(s2, breaks=50, add=T, col=rgb(0,0,1,1))
abline(v=mean(s1), col="green", lwd=3)
# hist(s2, breaks=50, add=TRUE, col = rgb(0, 0, 1, 0.5))
abline(v=mean(s2), col="lightblue", lwd=3)
diff <- mean(s1)-mean(s2)
se.ts
diff/se.ts
####
# repeated measure t-test
# we can use the above case
# pop paramter unknown
# two consecutive measurement
# for the same sample
t1 <- s1
t2 <- s2
mean(t1)
mean(t2)
diff.s <- t1 - t2
diff.s
t.cal.rm <- mean(diff.s)/se(diff.s)
t.cal.rm
p.val.rm <- pt(abs(t.cal.rm), length(s1)-1, lower.tail = F) * 2
p.val.rm
t.test(s1, s2, paired = T)
# create multiple histogram
s.all <- c(s1,s2)
mean(s.all)
hist(s1, col='grey', breaks=50, xlim=c(50, 150))
hist(s2, col='darkgreen', breaks=50, add=TRUE)
abline(v=c(mean(s.all)),
col=c("red"), lwd=3)
abline(v=c(mean(s1), mean(s2)),
col=c("black", "green"), lwd=3)
comb <- data.frame(s1,s2)
dat <- stack(comb)
head(dat)
m.tot <- mean(s.all)
m.s1 <- mean(s1)
m.s2 <- mean(s2)
ss.tot <- ss(s.all)
ss.s1 <- ss(s1)
ss.s2 <- ss(s2)
df.tot <- length(s.all)-1
df.s1 <- length(s1)-1
df.s2 <- length(s2)-1
ms.tot <- var(s.all)
ms.tot
ss.tot/df.tot
var(s1)
ss.s1 / df.s1
var(s2)
ss.s2 / df.s2
ss.b.s1 <- length(s1) * ((m.tot - m.s1)^2)
ss.b.s2 <- length(s2) * ((m.tot - m.s1)^2)
ss.bet <- ss.b.s1+ss.b.s2
ss.bet
ss.wit <- ss.s1 + ss.s2
ss.wit
ss.bet + ss.wit
ss.tot
library(dplyr)
# df.bet <- length(unique(dat)) - 1
df.bet <- nlevels(dat$ind) - 1
df.wit <- df.s1+df.s2
df.bet
df.wit
df.bet+df.wit
df.tot
ms.bet <- ss.bet / df.bet
ms.wit <- ss.wit / df.wit
ms.bet
ms.wit
f.cal <- ms.bet / ms.wit
f.cal
pf(f.cal, df.bet, df.wit, lower.tail = F)
f.test <- aov(dat$values~ dat$ind, data = dat)
summary(f.test)
sqrt(f.cal)
t.cal.ts
# this is anova after all.
note.w02.1758192015.txt.gz · Last modified: by hkimscil