rm(list=ls())
rnorm2 <- function(n,mean,sd){
mean+sd*scale(rnorm(n))
}
n.p <- 10000
m.p <- 100
sd.p <- 10
p1 <- rnorm2(n.p, m.p, sd.p)
m.p1 <- mean(p1)
sd.p1 <- sd(p1)
p2 <- rnorm2(n.p, m.p+5, sd.p)
m.p2 <- mean(p2)
sd.p2 <- sd(p2)
n.s <- 100
se.z <- c(sqrt(var(p1)/n.s))
x_values <- seq(mean(p1)-5*se.z,
mean(p1)+15*se.z,
length.out = 500)
# Calculate the probability
# density for a normal distribution
y_values <- dnorm(x_values,
mean = mean(p1),
sd = se.z)
# Plot the theoretical PDF
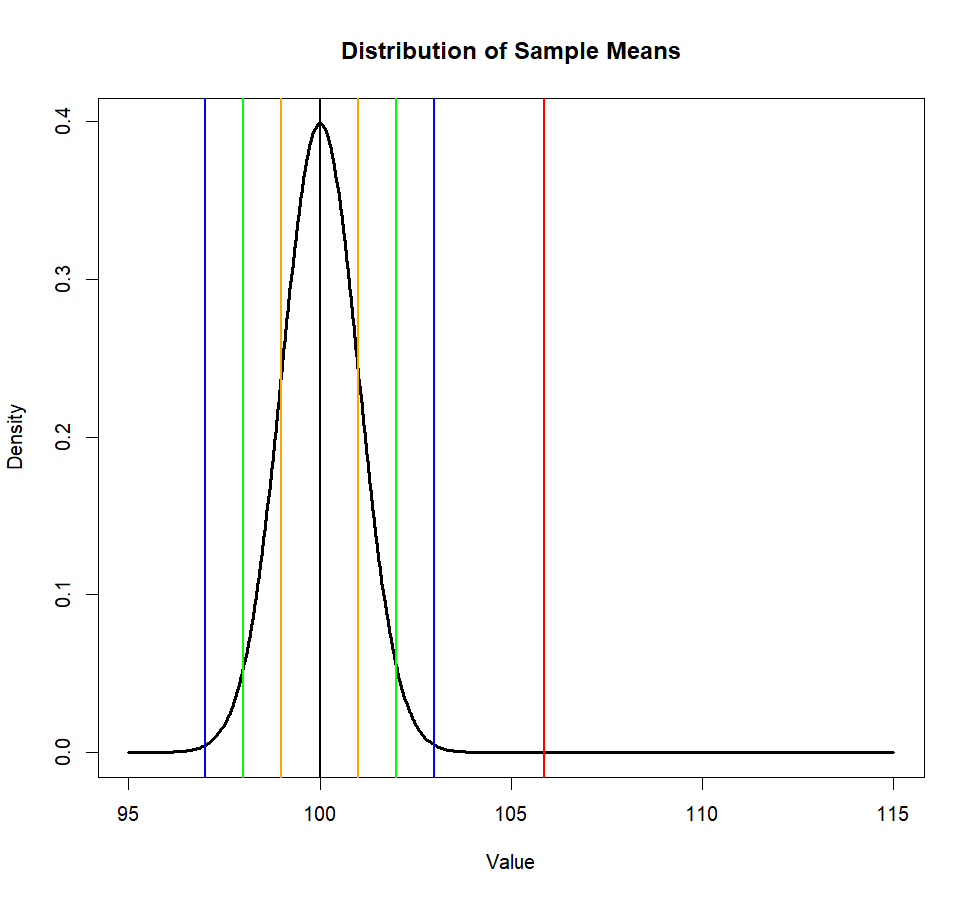
plot(x_values, y_values, type = "l",
lwd=3,
main = "Distribution of Sample Means",
xlab = "Value", ylab = "Density")
m.p1 <- mean(p1)
se1 <- c(m.p1-se.z, m.p1+se.z)
se2 <- c(m.p1-2*se.z, m.p1+2*se.z)
se3 <- c(m.p1-3*se.z, m.p1+3*se.z)
abline(v=c(m.p1,se1,se2,se3),
col=c('black', 'orange', 'orange',
'green', 'green',
'blue', 'blue'),
lwd=2)
treated.s <- sample(p2, n.s)
m.treated.s <- mean(treated.s)
abline(v=m.treated.s, col='red', lwd=2)
se.z
diff <- m.treated.s-mean(p1)
diff/se.z
# usual way - using sample's variance
# instead of p1's variance to get
# standard error value
se.s <- sqrt(var(treated.s)/n.s)
se.s
diff/se.s
pt(diff/se.s, df=n.s-1, lower.tail = F) * 2
t.test(treated.s, mu=m.p1, var.equal = T)
>
> rm(list=ls())
>
> rnorm2 <- function(n,mean,sd){
+ mean+sd*scale(rnorm(n))
+ }
>
> n.p <- 10000
> m.p <- 100
> sd.p <- 10
> p1 <- rnorm2(n.p, m.p, sd.p)
> m.p1 <- mean(p1)
> sd.p1 <- sd(p1)
>
> p2 <- rnorm2(n.p, m.p+5, sd.p)
> m.p2 <- mean(p2)
> sd.p2 <- sd(p2)
>
> n.s <- 100
> se.z <- c(sqrt(var(p1)/n.s))
>
> x_values <- seq(mean(p1)-5*se.z,
+ mean(p1)+15*se.z,
+ length.out = 500)
> # Calculate the probability
> # density for a normal distribution
> y_values <- dnorm(x_values,
+ mean = mean(p1),
+ sd = se.z)
>
> # Plot the theoretical PDF
> plot(x_values, y_values, type = "l",
+ lwd=3,
+ main = "Distribution of Sample Means",
+ xlab = "Value", ylab = "Density")
>
> m.p1 <- mean(p1)
> se1 <- c(m.p1-se.z, m.p1+se.z)
> se2 <- c(m.p1-2*se.z, m.p1+2*se.z)
> se3 <- c(m.p1-3*se.z, m.p1+3*se.z)
> abline(v=c(m.p1,se1,se2,se3),
+ col=c('black', 'orange', 'orange',
+ 'green', 'green',
+ 'blue', 'blue'),
+ lwd=2)
>
> treated.s <- sample(p2, n.s)
> m.treated.s <- mean(treated.s)
> abline(v=m.treated.s, col='red', lwd=2)
>
> se.z
[1] 1
>
> diff <- m.treated.s-mean(p1)
> diff/se.z
[1] 5.871217
>
> # usual way - using sample's variance
> # instead of p1's variance to get
> # standard error value
> se.s <- sqrt(var(treated.s)/n.s)
> se.s
[1] 0.9861042
> diff/se.s
[1] 5.953951
>
> pt(diff/se.s, df=n.s-1, lower.tail = F) * 2
[1] 3.994557e-08
> t.test(treated.s, mu=m.p1, var.equal = T)
One Sample t-test
data: treated.s
t = 5.954, df = 99, p-value = 3.995e-08
alternative hypothesis: true mean is not equal to 100
95 percent confidence interval:
103.9146 107.8279
sample estimates:
mean of x
105.8712
>