Table of Contents
t-test summing up
rm(list=ls())
rnorm2 <- function(n,mean,sd){
mean+sd*scale(rnorm(n))
}
se <- function(sample) {
sd(sample)/sqrt(length(sample))
}
ss <- function(x) {
sum((x-mean(x))^2)
}
N.p <- 100000
m.p <- 100
sd.p <- 10
p1 <- rnorm2(N.p, m.p, sd.p)
mean(p1)
sd(p1)
p2 <- rnorm2(N.p, m.p+10, sd.p)
mean(p2)
sd(p2)
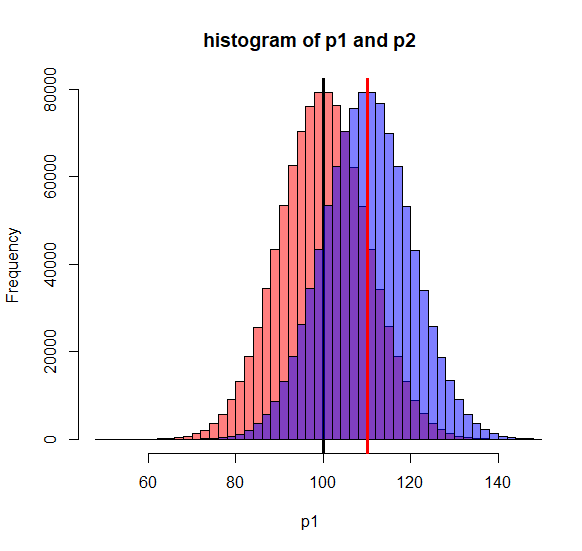
hist(p1, breaks=50, col = rgb(1, 0, 0, 0.5),
main = "histogram of p1 and p2",)
abline(v=mean(p1), col="black", lwd=3)
hist(p2, breaks=50, add=TRUE, col = rgb(0, 0, 1, 0.5))
abline(v=mean(p2), col="red", lwd=3)
# but suppose that we do not
# know the parameter of p2.
# But, the real effect of the
# drug is 10 point improvement
# (mean = 110).
hist(p1, breaks=50, col = rgb(1, 0, 0, 0.5),
main = "histogram of p1",)
abline(v=mean(p1), col="black", lwd=3)
# we take 100 random sample
# (from p2)
s.size <- 100
s2 <- sample(p2, s.size)
mean(s2)
sd(s2)
# The sample statistics are
# our clue to prove the
# effectiveness of the drug.
# Let's assume that the drug is
# NOT effective. And we also assume
# the distribution of sample means
# whose sample size is 100 (in this
# case).
se.z <- sqrt(var(p1)/s.size)
se.z <- c(se.z)
z.cal <- (mean(s2) - mean(p1)) / se.z
z.cal
pnorm(z.cal, lower.tail = F) * 2
# So,
iter <- 100000
means <- rep(NA, iter)
# means <- c()
for (i in 1:iter) {
m.of.s <- mean(sample(p1, s.size))
# means <- append(means, m.of.s)
means[i] <- m.of.s
}
hist(means,
xlim = c(mean(means)-3*sd(means),
mean(means)+10*sd(means)),
col = rgb(0, 0, 0, 0))
abline(v=mean(p1), col="black", lwd=3)
abline(v=mean(s2),
col="blue", lwd=3)
lo1 <- mean(p1)-se.z
hi1 <- mean(p1)+se.z
lo2 <- mean(p1)-2*se.z
hi2 <- mean(p1)+2*se.z
abline(v=c(lo1, hi1, lo2, hi2),
col=c("green","green", "brown", "brown"),
lwd=2)
se.z
c(lo2, hi2)
pnorm(z.cal, lower.tail = F) * 2
# Note that we use sqrt(var(s2)/s.size)
# as our se value instread of
# sqrt(var(p1)/s.size)
# This is a common practice for R
# In fact, some z.test (made by someone)
# function uses the former rather than
# latter.
sqrt(var(p1)/s.size)
se.z
sqrt(var(s2)/s.size)
se(s2)
se.s <- se(s2)
t.cal.os <- (mean(s2) - mean(p1))/ se.s
z.cal <- (mean(s2) - mean(p1)) / se.z
t.cal.os
z.cal
df.s2 <- length(s2) - 1
df.s2
p.t.os <- pt(abs(t.cal.os),
df.s2,
lower.tail = F) * 2
t.out <- t.test(s2, mu=mean(p1))
t.out
t.cal.os
p.t.os
########################
# 2 sample t-test
########################
# 가정. 아래에서 추출하는 두
# 샘플의 모집단의 파라미터를
# 모른다.
s1 <- sample(p1, s.size)
s2 <- sample(p2, s.size)
mean(s1)
mean(s2)
ss(s1)
ss(s2)
df.s1 <- length(s1)-1
df.s2 <- length(s2)-1
df.s1
df.s2
pooled.variance <- (ss(s1)+ss(s2))/(df.s1+df.s2)
pooled.variance
se.ts <- sqrt((pooled.variance/length(s1))+(pooled.variance/length(s2)))
se.ts
t.cal.ts <- (mean(s1)-mean(s2))/se.ts
t.cal.ts
p.val.ts <- pt(abs(t.cal.ts), df=df.s1+df.s2, lower.tail = F) * 2
p.val.ts
t.out <- t.test(s1, s2, var.equal = T)
t.out
hist(s1, breaks=30,
col = rgb(1, 0, 0, 0.5))
hist(s2, breaks=30, add=T, col=rgb(0,0,1,1))
abline(v=mean(s1), col="green", lwd=3)
# hist(s2, breaks=50, add=TRUE, col = rgb(0, 0, 1, 0.5))
abline(v=mean(s2), col="lightblue", lwd=3)
diff <- (mean(s1)-mean(s2))
diff
se.ts
diff/se.ts
t.out$statistic
####
# repeated measure t-test
# we can use the above case
# pop paramter unknown
# two consecutive measurement
# for the same sample
t1 <- s1
t2 <- s2
mean(t1)
mean(t2)
diff.s <- t1 - t2
head(diff.s)
t.cal.rm <- mean(diff.s)/se(diff.s)
t.cal.rm
p.val.rm <- pt(abs(t.cal.rm),
length(s1)-1,
lower.tail = F) * 2
p.val.rm
t.test(s1, s2, paired = T)
# 2 샘플 히스토그램을 이용해서
# anova 설명하기
s.all <- c(s1,s2)
mean(s.all) # 두 집단을 합친
# 종속변인의 평균 (아래 히스토
# 그램에서 검정색).
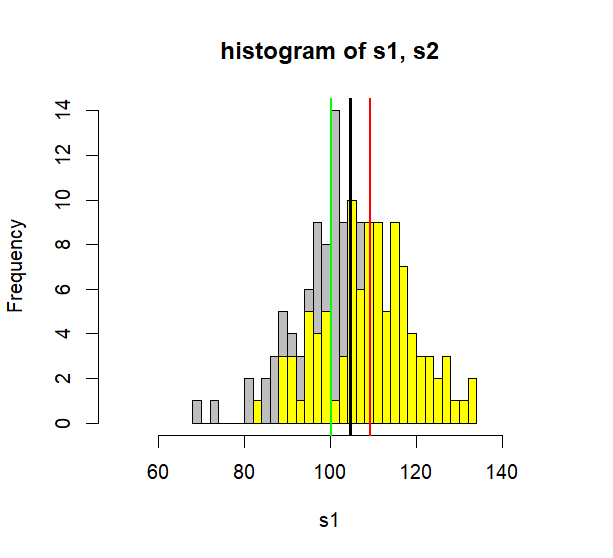
hist(s1, col='grey', breaks=30, xlim=c(50, 150),
main = "histogram of s1, s2")
hist(s2, col='yellow', breaks=30, add=TRUE)
abline(v=c(mean(s.all)),
col=c("black"), lwd=3)
abline(v=c(mean(s1), mean(s2)),
col=c("green", "red"), lwd=2)
comb <- data.frame(s1,s2)
dat <- stack(comb)
head(dat)
tail(dat)
m.tot <- mean(s.all)
m.s1 <- mean(s1)
m.s2 <- mean(s2)
ss.tot <- ss(s.all)
bet.s1 <- (m.tot - m.s1)^2 * length(s1)
bet.s2 <- (m.tot - m.s2)^2 * length(s2)
ss.bet <- bet.s1 + bet.s2
ss.s1 <- ss(s1)
ss.s2 <- ss(s2)
ss.wit <- ss.s1+ss.s2
ss.tot
ss.bet
ss.wit
ss.bet+ss.wit
df.tot <- length(s.all) - 1
df.bet <- nlevels(dat$ind) - 1
df.s1 <- length(s1)-1
df.s2 <- length(s2)-1
df.wit <- df.s1 + df.s2
df.tot
df.bet
df.wit
df.bet+df.wit
ss.tot/df.tot
ms.tot <- ss.tot/df.tot
ms.bet <- ss.bet / df.bet
ms.wit <- ss.wit / df.wit
f.cal <- ms.bet / ms.wit
f.cal
pf(f.cal, df.bet, df.wit, lower.tail = F)
f.test <- aov(dat$values~ dat$ind, data = dat)
sum.f.test <- summary(f.test)
sum.f.test
sum.f.test[[1]][1,4]
sum.f.test[[1]][1,5]
sqrt(f.cal)
t.cal.ts
# the above is anova after all.
m1 <- lm(dat$values~dat$ind, data = dat)
sum.m1 <- summary(m1)
sum.m1
sum.m1$r.square
ss.bet # 독립변인인 s1, s2의 그룹
# 구분으로 설명된 ss.tot 중의 일부
ss.tot # y 전체의 uncertainty
ss.bet/ss.tot
sum.m1$fstatistic[1]
ms.bet/ms.wit
t-test summing up output
1
> rm(list=ls())
> rnorm2 <- function(n,mean,sd){
+ mean+sd*scale(rnorm(n))
+ }
> se <- function(sample) {
+ sd(sample)/sqrt(length(sample))
+ }
> ss <- function(x) {
+ sum((x-mean(x))^2)
+ }
>
EXP.
- 필요한 펑션들
- se standard error 값을 구하는 펑션
- ss sum of square 값을 구하는 펑션
2
> N.p <- 100000 > m.p <- 100 > sd.p <- 10 > > > p1 <- rnorm2(N.p, m.p, sd.p) > mean(p1) [1] 100 > sd(p1) [1] 10 > > p2 <- rnorm2(N.p, m.p+10, sd.p) > mean(p2) [1] 110 > sd(p2) [1] 10 > > hist(p1, breaks=50, col = rgb(1, 0, 0, 0.5), + main = "histogram of p1 and p2",) > abline(v=mean(p1), col="black", lwd=3) > hist(p2, breaks=50, add=TRUE, col = rgb(0, 0, 1, 0.5)) > abline(v=mean(p2), col="red", lwd=3) > > # but suppose that we do not > # know the parameter of p2. > # But, the real effect of the > # drug is 10 point improvement > # (mean = 110). >
3
> # we take 100 random sample > # (from p2) > s.size <- 100 > s2 <- sample(p2, s.size) > mean(s2) [1] 109.5224 > sd(s2) [1] 9.996692 > > # The sample statistics are > # our clue to prove the > # effectiveness of the drug. > > # Let's assume that the drug is > # NOT effective. And we also assume > # the distribution of sample means > # whose sample size is 100 (in this > # case). > > se.z <- sqrt(var(p1)/s.size) > se.z <- c(se.z) > > z.cal <- (mean(s2) - mean(p1)) / se.z > z.cal [1] 9.522449 > pnorm(z.cal, lower.tail = F) * 2 [1] 1.691454e-21 >
…………………………..
4
> # So,
> iter <- 100000
> means <- rep(NA, iter)
> # means <- c()
> for (i in 1:iter) {
+ m.of.s <- mean(sample(p1, s.size))
+ # means <- append(means, m.of.s)
+ means[i] <- m.of.s
+ }
>
> hist(means,
+ xlim = c(mean(means)-3*sd(means),
+ mean(means)+10*sd(means)),
+ col = rgb(0, 0, 0, 0))
> abline(v=mean(p1), col="black", lwd=3)
> abline(v=mean(s2),
+ col="blue", lwd=3)
> lo1 <- mean(p1)-se.z
> hi1 <- mean(p1)+se.z
> lo2 <- mean(p1)-2*se.z
> hi2 <- mean(p1)+2*se.z
> abline(v=c(lo1, hi1, lo2, hi2),
+ col=c("green","green", "brown", "brown"),
+ lwd=2)
> se.z
[1] 1
> c(lo2, hi2)
[1] 98 102
> pnorm(z.cal, lower.tail = F) * 2
[1] 1.691454e-21
>
>
…………………………..
5
> # Note that we use sqrt(var(s2)/s.size)
> # as our se value instread of
> # sqrt(var(p1)/s.size)
> # This is a common practice for R
> # In fact, some z.test (made by someone)
> # function uses the former rather than
> # latter.
>
> sqrt(var(p1)/s.size)
[,1]
[1,] 1
> se.z
[1] 1
>
> sqrt(var(s2)/s.size)
[1] 0.9996692
> se(s2)
[1] 0.9996692
> se.s <- se(s2)
>
>
> t.cal.os <- (mean(s2) - mean(p1))/ se.s
> z.cal <- (mean(s2) - mean(p1)) / se.z
>
> t.cal.os
[1] 9.5256
> z.cal
[1] 9.522449
>
> df.s2 <- length(s2) - 1
> df.s2
[1] 99
> p.t.os <- pt(abs(t.cal.os),
+ df.s2,
+ lower.tail = F) * 2
>
> t.out <- t.test(s2, mu=mean(p1))
> t.out
One Sample t-test
data: s2
t = 9.5256, df = 99, p-value = 1.186e-15
alternative hypothesis: true mean is not equal to 100
95 percent confidence interval:
107.5389 111.5060
sample estimates:
mean of x
109.5224
> t.cal.os
[1] 9.5256
> p.t.os
[1] 1.185895e-15
>
…………………………..
6
> ########################
> # 2 sample t-test
> ########################
> # 가정. 아래에서 추출하는 두
> # 샘플의 모집단의 파라미터를
> # 모른다.
> s1 <- sample(p1, s.size)
> s2 <- sample(p2, s.size)
>
> mean(s1)
[1] 100.3577
> mean(s2)
[1] 110.1579
> ss(s1)
[1] 7831.618
> ss(s2)
[1] 7809.363
> df.s1 <- length(s1)-1
> df.s2 <- length(s2)-1
> df.s1
[1] 99
> df.s2
[1] 99
>
> pooled.variance <- (ss(s1)+ss(s2))/(df.s1+df.s2)
> pooled.variance
[1] 78.99486
> se.ts <- sqrt((pooled.variance/length(s1))+(pooled.variance/length(s2)))
> se.ts
[1] 1.25694
> t.cal.ts <- (mean(s1)-mean(s2))/se.ts
> t.cal.ts
[1] -7.79685
> p.val.ts <- pt(abs(t.cal.ts), df=df.s1+df.s2, lower.tail = F) * 2
> p.val.ts
[1] 3.537633e-13
>
> t.out <- t.test(s1, s2, var.equal = T)
> t.out
Two Sample t-test
data: s1 and s2
t = -7.7969, df = 198, p-value = 3.538e-13
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-12.278877 -7.321463
sample estimates:
mean of x mean of y
100.3577 110.1579
>
> hist(s1, breaks=30,
+ col = rgb(1, 0, 0, 0.5))
> hist(s2, breaks=30, add=T, col=rgb(0,0,1,1))
> abline(v=mean(s1), col="green", lwd=3)
> # hist(s2, breaks=50, add=TRUE, col = rgb(0, 0, 1, 0.5))
> abline(v=mean(s2), col="lightblue", lwd=3)
>
> diff <- (mean(s1)-mean(s2))
> diff
[1] -9.80017
> se.ts
[1] 1.25694
> diff/se.ts
[1] -7.79685
> t.out$statistic
t
-7.79685
>
…………………………..
7
> ####
> # repeated measure t-test
> # we can use the above case
> # pop paramter unknown
> # two consecutive measurement
> # for the same sample
>
> t1 <- s1
> t2 <- s2
> mean(t1)
[1] 100.3577
> mean(t2)
[1] 110.1579
> diff.s <- t1 - t2
> head(diff.s)
[1] -8.588426 -21.283258 -12.958503 -7.221480 -9.284982 -22.907759
> t.cal.rm <- mean(diff.s)/se(diff.s)
> t.cal.rm
[1] -8.215406
> p.val.rm <- pt(abs(t.cal.rm),
+ length(s1)-1,
+ lower.tail = F) * 2
> p.val.rm
[1] 8.276575e-13
> t.test(s1, s2, paired = T)
Paired t-test
data: s1 and s2
t = -8.2154, df = 99, p-value = 8.277e-13
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
-12.167145 -7.433195
sample estimates:
mean difference
-9.80017
>
…………………………..
8 t-test and anova
> # 2 샘플 히스토그램을 이용해서
> # anova 설명하기
> s.all <- c(s1,s2)
> mean(s.all) # 두 집단을 합친
[1] 104.7682
> # 종속변인의 평균 (아래 히스토
> # 그램에서 검정색).
> hist(s1, col='grey', breaks=30, xlim=c(50, 150),
+ main = "histogram of s1, s2")
> hist(s2, col='yellow', breaks=30, add=TRUE)
> abline(v=c(mean(s.all)),
+ col=c("black"), lwd=3)
> abline(v=c(mean(s1), mean(s2)),
+ col=c("green", "red"), lwd=2)
>
9
> comb <- data.frame(s1,s2)
> dat <- stack(comb)
> head(dat)
values ind
1 106.26706 s1
2 101.89180 s1
3 98.64593 s1
4 93.82977 s1
5 80.35249 s1
6 100.47300 s1
> tail(dat)
values ind
195 111.33398 s2
196 115.99752 s2
197 109.85412 s2
198 126.80207 s2
199 99.13395 s2
200 130.01417 s2
>
……………………………….
10
> m.tot <- mean(s.all) > m.s1 <- mean(s1) > m.s2 <- mean(s2) > > ss.tot <- ss(s.all) > bet.s1 <- (m.tot - m.s1)^2 * length(s1) > bet.s2 <- (m.tot - m.s2)^2 * length(s2) > ss.bet <- bet.s1 + bet.s2 > ss.s1 <- ss(s1) > ss.s2 <- ss(s2) > ss.wit <- ss.s1+ss.s2 > > ss.tot [1] 25203.89 > ss.bet [1] 4219.463 > ss.wit [1] 20984.43 > ss.bet+ss.wit [1] 25203.89 > > df.tot <- length(s.all) - 1 > df.bet <- nlevels(dat$ind) - 1 > df.s1 <- length(s1)-1 > df.s2 <- length(s2)-1 > df.wit <- df.s1 + df.s2 > > df.tot [1] 199 > df.bet [1] 1 > df.wit [1] 198 > df.bet+df.wit [1] 199 >
- ss.between 은 두개의 집단과 그 집단의 평균으로 이루어진 분산에서의 ss값을 말한다.
- 그림에서 녹색선 위에 s1의 원소들이 모여있고
- 붉은색 선위에 s2원소들이 모여 있다고치면
- 이 분산은 그룹간 분산이 (between group variance) 된다.
- 이를 구하기 위해서 ss.bet의 공식을 사용한다.
- ss.within의 경우에는 간단히 s1과 s2 집단의 ss값을 구하는 것으로
- 스크립트 초반에 언급된 ss값을 이용해서 구한후 더한다.
- df.between은 그룹의 숫자에서 1을 뺀 숫자
- df.within은 각 그룹내의 구성원에서 1을 뺀 후, 모두 더한 숫자이다
- df.total은 전체 구성원에서 1을 빼거나, df.bet + df.wit 를 더한 숫자
11
> ss.tot/df.tot
[1] 126.6527
> ms.tot <- ss.tot/df.tot
>
> ms.bet <- ss.bet / df.bet
> ms.wit <- ss.wit / df.wit
>
> f.cal <- ms.bet / ms.wit
> f.cal
[1] 39.81302
> pf(f.cal, df.bet, df.wit, lower.tail = F)
[1] 1.792613e-09
>
> f.test <- aov(dat$values~ dat$ind, data = dat)
> sum.f.test <- summary(f.test)
> sum.f.test
Df Sum Sq Mean Sq F value Pr(>F)
dat$ind 1 4219 4219 39.81 1.79e-09 ***
Residuals 198 20984 106
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
>
> sum.f.test[[1]][1,4]
[1] 39.81302
> sum.f.test[[1]][1,5]
[1] 1.792613e-09
> sqrt(f.cal)
[1] 6.309756
> t.cal.ts
[1] -6.309756
>
> # the above is anova after all.
>
………………………..
- 위에서 구한 ss 값들을 가지고 분산값을 구할 수 있다 (ms = mean square = variance)
- ms.between은 ss.bet/df.bet 값을 말하는 것으로 그룹이 나누어짐으로써 구할 수 있었던 분산이다. 이 부분은 그룹 때문에 생긴 (treatment때문에 생긴) 분산이다.
- ms.within은 ss.wit/df.wit 값을 말하는 것으로 그룹의 구성 때문에 구할 수 있었던 ss.between 를 제외한 나머지 ss값을 df.wit값으로 나누어준 값을 말한다. 이 부분이 random 파트이다.
- 이 둘의 비율을 F값이라고 하고, 이는 기본적으로
- group difference 를 random difference 로 나눈 값을 말하고 이는
- t-test의 difference/random error와 같은 형태를 갖는다
- 따라서, 분산을 이용해서 구한 F값은 표준편차를 이용해서 구한 t값의 제곱값을 갖게 된다.
12
> m1 <- lm(dat$values~dat$ind, data = dat)
> sum.m1 <- summary(m1)
> sum.m1
Call:
lm(formula = dat$values ~ dat$ind, data = dat)
Residuals:
Min 1Q Median 3Q Max
-31.4177 -5.7058 0.4976 6.2952 29.2586
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 100.175 1.029 97.31 < 2e-16 ***
dat$inds2 9.186 1.456 6.31 1.79e-09 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 10.29 on 198 degrees of freedom
Multiple R-squared: 0.1674, Adjusted R-squared: 0.1632
F-statistic: 39.81 on 1 and 198 DF, p-value: 1.793e-09
>
> sum.m1$r.square
[1] 0.1674131
> ss.bet # 독립변인인 s1, s2의 그룹
[1] 4219.463
> # 구분으로 설명된 ss.tot 중의 일부
> ss.tot # y 전체의 uncertainty
[1] 25203.89
> ss.bet/ss.tot
[1] 0.1674131
>
> sum.m1$fstatistic[1]
value
39.81302
> ms.bet/ms.wit
[1] 39.81302
>
>
…………………………..
- ss.tot 은 s1그룹과 s2그룹의 값을 한줄에 배열하여 구하는 종속변인의 (dat$values) ss값을 말한다. 즉, 독립변이 없을 때의 종속변인의 uncertainty를 말한다.
- ss.bet은 독립변인이 고려됨으로써 설명된 종속부분의 일부를 말한다.
- ss.tot = ss.bet + ss.wit
- total uncertainty (a) = treatment effect (b) + random effect
- b/a를 R square값이라고 부른다.