r:logistic_regression_analysis
This is an old revision of the document!
Table of Contents
Logitistic Regression Analysis
\begin{align*} \displaystyle ln \left( {\frac{p}{(1-p)}} \right) = a + bX \end{align*}
- p = 변인 X가 A일 확률
- 1-p = 변인 X가 A가 아닐 확률
- ln 은 e를 밑으로 하는 log 를 말한다
- $ln \left( {\frac{p}{(1-p)}} \right) $ 을 $\text{logit(p)}$ 로 부른다
\begin{align*} \text{logit(p)} & = ln \left( {\frac{p}{(1-p)}} \right) = a + bX \\ \frac{p}{1-p} & = e^{a+bX} \\ p & = e^{a+bX} * (1-p) \\ p & = e^{a+bX} - p * \left(e^{a+bX} \right) \\ p + p * \left(e^{a+bX} \right) & = e^{a+bX} \\ p * \left(1 + e^{a+bX} \right) & = e^{a+bX} \\ p & = \frac {e^{a+bX}} { \left(1 + e^{a+bX} \right)} \\ \end{align*}
- 위에서 계수 b값이 충분히 커서 X 가 커지면 p 값은 1로 수렴하고
- b값이 충분히 작아서 X가 아주 작아지면 p 값은 0에 가까이 간다
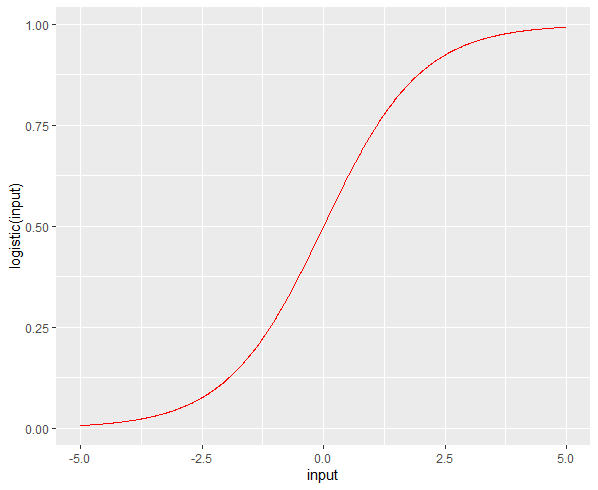
즉 p의 그래프는 아래와 같은 그래프의 곡선이다.
install.packages("sigmoid")
library(sigmoid)
library(ggplot2)
input <- seq(-5, 5, 0.01)
df = data.frame(input, logistic(input), Gompertz(input))
ggplot( df, aes(input, logistic(input)) ) +
geom_line(color="red")
ln 성질
여기서
\begin{align*}
y & = ln(x) \\
& = log_e {x} \\
x & = e^{y} \\
\end{align*}
위에서
- $ \text{if } \;\;\; x = 1, $
- $ e^{y} = 1 $ 이므로
- $ y = 0 $
- $ log_{e}(1) = 0 $
- $\text{if } \;\;\; x = 0 $
- $ 0 = e^{y} $ 이므로
- y 는 $ - \infty $
- 왜냐하면, $ e^{-\infty} = \frac {1}{e^{\infty}} = \frac {1}{\infty} = 0 $ 혹은 $0$ 에 수렴하기 때문
- $ log_{e}(0) = - \infty $
- $\text{if } \;\;\; x = \infty $
- $ \infty = e^{y} $ 이므로
- $ y = \infty $ 어야 함
- 따라서 $log_{e}(\infty) = + \infty $
e.g.
d <- subset(iris, Species == "virginica" | Species == "versicolor") head(d) d$Species <- factor(d$Species) str(d) m <- glm(Species ~ ., data=d, family="binomial") round(fitted(m)[c(1:5, 51:55)],3) round(fitted(m)[c(1:5, 51:55)],2) f <- fitted(m) as.numeric(d$Species) ifelse(f > .5, 1, 0) == as.numeric(d$Species) - 1 is_correct <- ifelse(f > .5, 1, 0) == as.numeric(d$Species) - 1 sum(is_correct) sum(is_correct) / NROW(is_correct) predict(m, newdata=d[c(1,10,55),], type="response") d3 <- read.csv(file="d3.csv") round(predict(m, newdata=d3[c(1:5),], type="response"),2) is_correct <- ifelse(f > .5, 1, 0) == as.numeric(d$Species) - 1 sum(is_correct) sum(is_correct) / NROW(is_correct)
> d <- subset(iris, Species == "virginica" | Species == "versicolor")
> d
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
51 7.0 3.2 4.7 1.4 versicolor
52 6.4 3.2 4.5 1.5 versicolor
53 6.9 3.1 4.9 1.5 versicolor
54 5.5 2.3 4.0 1.3 versicolor
55 6.5 2.8 4.6 1.5 versicolor
56 5.7 2.8 4.5 1.3 versicolor
57 6.3 3.3 4.7 1.6 versicolor
58 4.9 2.4 3.3 1.0 versicolor
59 6.6 2.9 4.6 1.3 versicolor
60 5.2 2.7 3.9 1.4 versicolor
61 5.0 2.0 3.5 1.0 versicolor
62 5.9 3.0 4.2 1.5 versicolor
63 6.0 2.2 4.0 1.0 versicolor
64 6.1 2.9 4.7 1.4 versicolor
65 5.6 2.9 3.6 1.3 versicolor
66 6.7 3.1 4.4 1.4 versicolor
67 5.6 3.0 4.5 1.5 versicolor
68 5.8 2.7 4.1 1.0 versicolor
69 6.2 2.2 4.5 1.5 versicolor
70 5.6 2.5 3.9 1.1 versicolor
71 5.9 3.2 4.8 1.8 versicolor
72 6.1 2.8 4.0 1.3 versicolor
73 6.3 2.5 4.9 1.5 versicolor
74 6.1 2.8 4.7 1.2 versicolor
75 6.4 2.9 4.3 1.3 versicolor
76 6.6 3.0 4.4 1.4 versicolor
77 6.8 2.8 4.8 1.4 versicolor
78 6.7 3.0 5.0 1.7 versicolor
79 6.0 2.9 4.5 1.5 versicolor
80 5.7 2.6 3.5 1.0 versicolor
81 5.5 2.4 3.8 1.1 versicolor
82 5.5 2.4 3.7 1.0 versicolor
83 5.8 2.7 3.9 1.2 versicolor
84 6.0 2.7 5.1 1.6 versicolor
85 5.4 3.0 4.5 1.5 versicolor
86 6.0 3.4 4.5 1.6 versicolor
87 6.7 3.1 4.7 1.5 versicolor
88 6.3 2.3 4.4 1.3 versicolor
89 5.6 3.0 4.1 1.3 versicolor
90 5.5 2.5 4.0 1.3 versicolor
91 5.5 2.6 4.4 1.2 versicolor
92 6.1 3.0 4.6 1.4 versicolor
93 5.8 2.6 4.0 1.2 versicolor
94 5.0 2.3 3.3 1.0 versicolor
95 5.6 2.7 4.2 1.3 versicolor
96 5.7 3.0 4.2 1.2 versicolor
97 5.7 2.9 4.2 1.3 versicolor
98 6.2 2.9 4.3 1.3 versicolor
99 5.1 2.5 3.0 1.1 versicolor
100 5.7 2.8 4.1 1.3 versicolor
101 6.3 3.3 6.0 2.5 virginica
102 5.8 2.7 5.1 1.9 virginica
103 7.1 3.0 5.9 2.1 virginica
104 6.3 2.9 5.6 1.8 virginica
105 6.5 3.0 5.8 2.2 virginica
106 7.6 3.0 6.6 2.1 virginica
107 4.9 2.5 4.5 1.7 virginica
108 7.3 2.9 6.3 1.8 virginica
109 6.7 2.5 5.8 1.8 virginica
110 7.2 3.6 6.1 2.5 virginica
111 6.5 3.2 5.1 2.0 virginica
112 6.4 2.7 5.3 1.9 virginica
113 6.8 3.0 5.5 2.1 virginica
114 5.7 2.5 5.0 2.0 virginica
115 5.8 2.8 5.1 2.4 virginica
116 6.4 3.2 5.3 2.3 virginica
117 6.5 3.0 5.5 1.8 virginica
118 7.7 3.8 6.7 2.2 virginica
119 7.7 2.6 6.9 2.3 virginica
120 6.0 2.2 5.0 1.5 virginica
121 6.9 3.2 5.7 2.3 virginica
122 5.6 2.8 4.9 2.0 virginica
123 7.7 2.8 6.7 2.0 virginica
124 6.3 2.7 4.9 1.8 virginica
125 6.7 3.3 5.7 2.1 virginica
126 7.2 3.2 6.0 1.8 virginica
127 6.2 2.8 4.8 1.8 virginica
128 6.1 3.0 4.9 1.8 virginica
129 6.4 2.8 5.6 2.1 virginica
130 7.2 3.0 5.8 1.6 virginica
131 7.4 2.8 6.1 1.9 virginica
132 7.9 3.8 6.4 2.0 virginica
133 6.4 2.8 5.6 2.2 virginica
134 6.3 2.8 5.1 1.5 virginica
135 6.1 2.6 5.6 1.4 virginica
136 7.7 3.0 6.1 2.3 virginica
137 6.3 3.4 5.6 2.4 virginica
138 6.4 3.1 5.5 1.8 virginica
139 6.0 3.0 4.8 1.8 virginica
140 6.9 3.1 5.4 2.1 virginica
141 6.7 3.1 5.6 2.4 virginica
142 6.9 3.1 5.1 2.3 virginica
143 5.8 2.7 5.1 1.9 virginica
144 6.8 3.2 5.9 2.3 virginica
145 6.7 3.3 5.7 2.5 virginica
146 6.7 3.0 5.2 2.3 virginica
147 6.3 2.5 5.0 1.9 virginica
148 6.5 3.0 5.2 2.0 virginica
149 6.2 3.4 5.4 2.3 virginica
150 5.9 3.0 5.1 1.8 virginica
> d$Species <- factor(d$Species) > str(d) 'data.frame': 100 obs. of 5 variables: $ Sepal.Length: num 7 6.4 6.9 5.5 6.5 5.7 6.3 4.9 6.6 5.2 ... $ Sepal.Width : num 3.2 3.2 3.1 2.3 2.8 2.8 3.3 2.4 2.9 2.7 ... $ Petal.Length: num 4.7 4.5 4.9 4 4.6 4.5 4.7 3.3 4.6 3.9 ... $ Petal.Width : num 1.4 1.5 1.5 1.3 1.5 1.3 1.6 1 1.3 1.4 ... $ Species : Factor w/ 2 levels "versicolor","virginica": 1 1 1 1 1 1 1 1 1 1 ...
> m <- glm(Species ~ ., data=d, family="binomial")
> m
Call: glm(formula = Species ~ ., family = "binomial", data = d)
Coefficients:
(Intercept) Sepal.Length Sepal.Width Petal.Length
-42.638 -2.465 -6.681 9.429
Petal.Width
18.286
Degrees of Freedom: 99 Total (i.e. Null); 95 Residual
Null Deviance: 138.6
Residual Deviance: 11.9 AIC: 21.9
> round(fitted(m)[c(1:5, 51:55)],3) 51 52 53 54 55 101 102 103 104 105 0.000 0.000 0.001 0.000 0.001 1.000 1.000 1.000 1.000 1.000 > round(fitted(m)[c(1:5, 51:55)],2) 51 52 53 54 55 101 102 103 104 105 0 0 0 0 0 1 1 1 1 1
> f <- fitted(m) > as.numeric(d$Species) [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 [32] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 [63] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 [94] 2 2 2 2 2 2 2 > ifelse(f > .5, 1, 0) == as.numeric(d$Species) - 1 51 52 53 54 55 56 57 58 59 60 61 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 62 63 64 65 66 67 68 69 70 71 72 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 73 74 75 76 77 78 79 80 81 82 83 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 84 85 86 87 88 89 90 91 92 93 94 FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 95 96 97 98 99 100 101 102 103 104 105 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 106 107 108 109 110 111 112 113 114 115 116 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 117 118 119 120 121 122 123 124 125 126 127 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 128 129 130 131 132 133 134 135 136 137 138 TRUE TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE 139 140 141 142 143 144 145 146 147 148 149 TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE 150 TRUE
> predict(m, newdata=d[c(1,10,55),], type="response")
51 60 105
1.171672e-05 1.481064e-05 9.999999e-01
> d3 <- read.csv("http://commres.net/wiki/_media/d3.csv")
> round(predict(m, newdata=d3[c(1:5),], type="response"),2)
1 2 3 4 5
1.00 1.00 0.92 0.00 0.00
r/logistic_regression_analysis.1701678303.txt.gz · Last modified: by hkimscil