r:twoway_repeated_measure_anova
see
E.g. 1
# rep.meas.anova.two.way.csv
df <- read.csv("http://commres.net/wiki/_media/r/rep.meas.anova.two.way.csv")
head(df)
tail(df)
str(df)
df$group <- factor(df$group)
df$id<-1:196
df$id <- factor(df$id)
str(df)
library(reshape2)
mltdf <- melt(df, id.var=c("id", "group"))
mltdf
colnames(mltdf) <- c("id", "group", "time", "value")
mltdf.ftest <- aov(value ~ group * time + Error(id), data=mltdf)
summary(mltdf.ftest)
attach(mltdf)
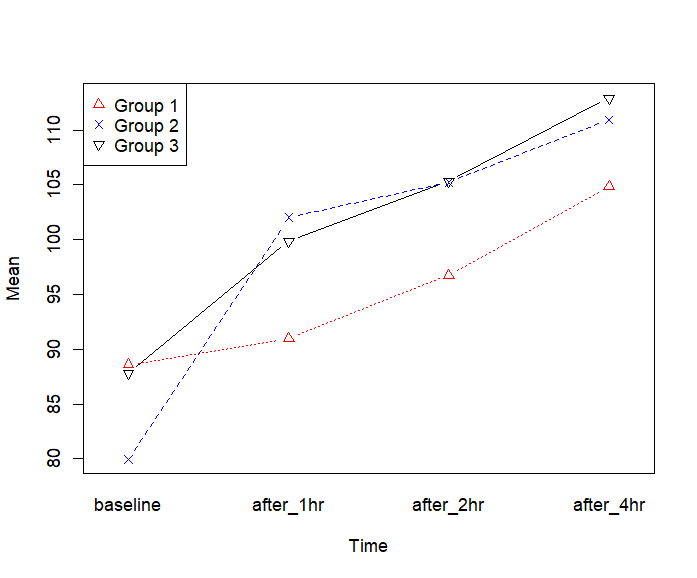
interaction.plot(time, group, value,
fun=mean, type="b", pch=c(2,4,6),
legend = F, col=c("red","blue","black"),
xlab="Time", ylab="Mean")
legend("topleft", legend=c("Group 1", "Group 2", "Group 3"),
pch=c(2,4,6), col=c("red", "blue", "black"))
pairwise.t.test(value, time,
paired=T,
p.adjust.method="bonferroni")
> # rep.meas.anova.two.way.csv
>
> df <- read.csv("http://commres.net/wiki/_media/r/rep.meas.anova.two.way.csv")
> head(df)
group baseline after_1hr after_2hr after_4hr
1 1 39.1 90.7 67.4 59.2
2 3 73.4 142.9 56.2 119.6
3 3 93.6 101.8 62.4 115.2
4 2 113.5 109.5 100.2 135.8
5 1 94.2 111.3 111.3 105.8
6 1 58.0 102.3 129.8 80.3
> tail(df)
group baseline after_1hr after_2hr after_4hr
191 1 71.3 141.3 120.5 65.0
192 3 38.9 116.8 120.3 102.6
193 2 37.7 93.8 91.6 89.9
194 1 133.8 95.9 103.2 111.4
195 2 137.9 132.7 140.5 66.0
196 1 125.1 43.8 112.6 78.6
> str(df)
'data.frame': 196 obs. of 5 variables:
$ group : int 1 3 3 2 1 1 2 1 2 1 ...
$ baseline : num 39.1 73.4 93.6 113.5 94.2 ...
$ after_1hr: num 90.7 142.9 101.8 109.5 111.3 ...
$ after_2hr: num 67.4 56.2 62.4 100.2 111.3 ...
$ after_4hr: num 59.2 119.6 115.2 135.8 105.8 ...
>
> df$group <- factor(df$group)
> df$id<-1:196
> df$id <- factor(df$id)
> str(df)
'data.frame': 196 obs. of 6 variables:
$ group : Factor w/ 3 levels "1","2","3": 1 3 3 2 1 1 2 1 2 1 ...
$ baseline : num 39.1 73.4 93.6 113.5 94.2 ...
$ after_1hr: num 90.7 142.9 101.8 109.5 111.3 ...
$ after_2hr: num 67.4 56.2 62.4 100.2 111.3 ...
$ after_4hr: num 59.2 119.6 115.2 135.8 105.8 ...
$ id : Factor w/ 196 levels "1","2","3","4",..: 1 2 3 4 5 6 7 8 9 10 ...
>
> library(reshape2)
> mltdf <- melt(df, id.var=c("id", "group"))
> mltdf
id group variable value
1 1 1 baseline 39.1
2 2 3 baseline 73.4
3 3 3 baseline 93.6
4 4 2 baseline 113.5
5 5 1 baseline 94.2
6 6 1 baseline 58.0
7 7 2 baseline 90.2
8 8 1 baseline 94.0
9 9 2 baseline 52.4
10 10 1 baseline 61.6
11 11 2 baseline 116.1
12 12 1 baseline 95.8
13 13 2 baseline 130.7
14 14 3 baseline 34.6
15 15 2 baseline 72.9
16 16 1 baseline 64.5
17 17 2 baseline 76.1
18 18 1 baseline 118.6
19 19 2 baseline 46.5
20 20 3 baseline 124.6
21 21 3 baseline 50.3
22 22 1 baseline 67.7
23 23 1 baseline 100.5
24 24 1 baseline 66.4
25 25 2 baseline 129.7
26 26 1 baseline 92.2
27 27 1 baseline 49.1
28 28 2 baseline 89.3
29 29 1 baseline 101.5
30 30 1 baseline 110.5
31 31 1 baseline 95.4
32 32 1 baseline 59.6
33 33 2 baseline 87.6
34 34 3 baseline 42.3
35 35 3 baseline 110.0
36 36 3 baseline 133.6
37 37 2 baseline 36.8
38 38 1 baseline 106.9
39 39 3 baseline 32.1
40 40 1 baseline 118.9
41 41 2 baseline 70.3
42 42 1 baseline 105.2
43 43 1 baseline 55.2
44 44 3 baseline 103.4
45 45 2 baseline 55.9
46 46 2 baseline 93.3
47 47 3 baseline 113.0
48 48 2 baseline 44.8
49 49 1 baseline 125.4
50 50 2 baseline 91.5
51 51 2 baseline 46.8
52 52 3 baseline 46.9
53 53 1 baseline 88.4
54 54 1 baseline 83.1
55 55 1 baseline 72.7
56 56 1 baseline 78.3
57 57 1 baseline 73.1
58 58 1 baseline 69.9
59 59 2 baseline 94.2
60 60 3 baseline 133.1
61 61 1 baseline 69.7
62 62 1 baseline 95.9
63 63 1 baseline 122.1
64 64 2 baseline 46.4
65 65 3 baseline 62.3
66 66 2 baseline 134.0
67 67 2 baseline 66.9
68 68 3 baseline 109.1
69 69 1 baseline 100.1
70 70 3 baseline 54.9
71 71 2 baseline 118.9
72 72 2 baseline 79.7
73 73 1 baseline 85.2
74 74 3 baseline 133.5
75 75 1 baseline 125.6
76 76 2 baseline 70.3
77 77 3 baseline 133.0
78 78 2 baseline 132.7
79 79 3 baseline 134.6
80 80 2 baseline 97.2
81 81 1 baseline 70.9
82 82 1 baseline 99.3
83 83 3 baseline 131.4
84 84 3 baseline 88.9
85 85 2 baseline 77.2
86 86 1 baseline 62.7
87 87 1 baseline 49.0
88 88 3 baseline 125.1
89 89 3 baseline 94.8
90 90 3 baseline 61.0
91 91 1 baseline 132.9
92 92 2 baseline 109.5
93 93 3 baseline 53.6
94 94 2 baseline 80.4
95 95 2 baseline 48.5
96 96 1 baseline 129.1
97 97 2 baseline 119.3
98 98 2 baseline 74.1
99 99 2 baseline 59.2
100 100 2 baseline 115.5
101 101 2 baseline 125.4
102 102 2 baseline 111.0
103 103 2 baseline 63.5
104 104 2 baseline 60.8
105 105 3 baseline 107.9
106 106 3 baseline 69.2
107 107 3 baseline 133.7
108 108 3 baseline 45.1
109 109 3 baseline 108.4
110 110 2 baseline 69.6
111 111 2 baseline 94.0
112 112 3 baseline 34.2
113 113 2 baseline 41.6
114 114 1 baseline 46.7
115 115 3 baseline 97.2
116 116 1 baseline 70.5
117 117 1 baseline 97.9
118 118 1 baseline 39.6
119 119 2 baseline 39.1
120 120 2 baseline 128.9
121 121 3 baseline 132.5
122 122 3 baseline 71.3
123 123 3 baseline 130.5
124 124 1 baseline 106.9
125 125 2 baseline 84.5
126 126 1 baseline 118.1
127 127 2 baseline 38.0
128 128 3 baseline 51.2
129 129 2 baseline 107.8
130 130 1 baseline 56.8
131 131 2 baseline 71.9
132 132 2 baseline 90.1
133 133 2 baseline 81.4
134 134 3 baseline 88.5
135 135 3 baseline 119.5
136 136 2 baseline 58.2
137 137 1 baseline 109.0
138 138 3 baseline 97.0
139 139 1 baseline 132.0
140 140 2 baseline 35.8
141 141 1 baseline 96.0
142 142 1 baseline 35.9
143 143 1 baseline 65.2
144 144 3 baseline 121.0
145 145 2 baseline 122.8
146 146 2 baseline 51.1
147 147 2 baseline 51.5
148 148 1 baseline 108.7
149 149 1 baseline 89.5
150 150 1 baseline 86.8
151 151 3 baseline 49.2
152 152 3 baseline 119.1
153 153 1 baseline 86.7
154 154 2 baseline 39.9
155 155 1 baseline 46.9
156 156 3 baseline 64.5
157 157 2 baseline 39.1
158 158 1 baseline 70.9
159 159 2 baseline 84.0
160 160 3 baseline 105.2
161 161 2 baseline 46.0
162 162 3 baseline 98.6
163 163 1 baseline 87.3
164 164 2 baseline 52.5
165 165 1 baseline 125.4
166 166 1 baseline 73.7
167 167 1 baseline 101.2
168 168 2 baseline 97.1
169 169 1 baseline 44.9
170 170 1 baseline 39.1
171 171 1 baseline 80.3
172 172 2 baseline 107.5
173 173 3 baseline 67.6
174 174 3 baseline 126.5
175 175 1 baseline 97.8
176 176 1 baseline 132.4
177 177 2 baseline 39.3
178 178 3 baseline 36.1
179 179 3 baseline 30.6
180 180 1 baseline 110.8
181 181 1 baseline 124.5
182 182 3 baseline 78.5
183 183 1 baseline 110.0
184 184 3 baseline 72.2
185 185 1 baseline 121.1
186 186 1 baseline 47.8
187 187 1 baseline 125.6
188 188 1 baseline 116.2
189 189 2 baseline 85.7
190 190 2 baseline 94.5
191 191 1 baseline 71.3
192 192 3 baseline 38.9
193 193 2 baseline 37.7
194 194 1 baseline 133.8
195 195 2 baseline 137.9
196 196 1 baseline 125.1
197 1 1 after_1hr 90.7
198 2 3 after_1hr 142.9
199 3 3 after_1hr 101.8
200 4 2 after_1hr 109.5
201 5 1 after_1hr 111.3
202 6 1 after_1hr 102.3
203 7 2 after_1hr 112.4
204 8 1 after_1hr 76.8
205 9 2 after_1hr 147.1
206 10 1 after_1hr 88.9
207 11 2 after_1hr 69.7
208 12 1 after_1hr 127.5
209 13 2 after_1hr 108.3
210 14 3 after_1hr 107.1
211 15 2 after_1hr 112.6
212 16 1 after_1hr 61.5
213 17 2 after_1hr 118.2
214 18 1 after_1hr 108.8
215 19 2 after_1hr 136.3
216 20 3 after_1hr 91.7
217 21 3 after_1hr 79.0
218 22 1 after_1hr 55.7
219 23 1 after_1hr 67.3
220 24 1 after_1hr 86.5
221 25 2 after_1hr 123.9
222 26 1 after_1hr 126.2
223 27 1 after_1hr 41.3
224 28 2 after_1hr 62.1
225 29 1 after_1hr 102.6
226 30 1 after_1hr 62.9
227 31 1 after_1hr 68.9
228 32 1 after_1hr 77.8
229 33 2 after_1hr 92.8
230 34 3 after_1hr 56.8
231 35 3 after_1hr 87.0
232 36 3 after_1hr 126.3
233 37 2 after_1hr 115.2
234 38 1 after_1hr 80.5
235 39 3 after_1hr 75.9
236 40 1 after_1hr 85.5
237 41 2 after_1hr 125.5
238 42 1 after_1hr 93.6
239 43 1 after_1hr 99.9
240 44 3 after_1hr 70.8
241 45 2 after_1hr 111.9
242 46 2 after_1hr 60.5
243 47 3 after_1hr 132.6
244 48 2 after_1hr 137.5
245 49 1 after_1hr 95.9
246 50 2 after_1hr 59.0
247 51 2 after_1hr 62.5
248 52 3 after_1hr 138.2
249 53 1 after_1hr 40.5
250 54 1 after_1hr 55.7
[ reached 'max' / getOption("max.print") -- omitted 534 rows ]
> colnames(mltdf) <- c("id", "group", "time", "value")
>
> mltdf.ftest <- aov(value ~ group * time + Error(id), data=mltdf)
> summary(mltdf.ftest)
Error: id
Df Sum Sq Mean Sq F value Pr(>F)
group 2 5283 2641.6 3.298 0.0391 *
Residuals 193 154611 801.1
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
time 3 58104 19368 20.747 8.85e-13 ***
group:time 6 8427 1405 1.505 0.174
Residuals 579 540521 934
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
> attach(mltdf)
The following objects are masked from mltdf (pos = 4):
group, id, time, value
> interaction.plot(time, group, value,
+ fun=mean, type="b", pch=c(2,4,6),
+ legend = F, col=c("red","blue","black"),
+ xlab="Time", ylab="Mean")
> legend("topleft", legend=c("Group 1", "Group 2", "Group 3"),
+ pch=c(2,4,6), col=c("red", "blue", "black"))
>
> pairwise.t.test(value, time,
+ paired=T,
+ p.adjust.method="bonferroni")
Pairwise comparisons using paired t tests
data: value and time
baseline after_1hr after_2hr
after_1hr 0.0008 - -
after_2hr 2.5e-06 0.7653 -
after_4hr 4.7e-13 0.0026 0.1005
P value adjustment method: bonferroni
>
E.g. 2
acne <- read.csv("http://commres.net/wiki/_media/r/10_rmanova.csv")
str(acne)
acne
# install.packages("reshape")
library(reshape)
acne.re <- reshape(acne, direction="long", varying=3:6, sep="")
str(acne.re)
acne.re$group <- factor(acne.re$group)
acne.re$id <- factor(acne.re$id)
acne.re$time <- factor(acne.re$time)
str(acne.re)
acne.re
attach(acne.re)
acne.re.anova <- aov(month~group*time+Error(id/time), data=acne.re)
summary(acne.re.anova)
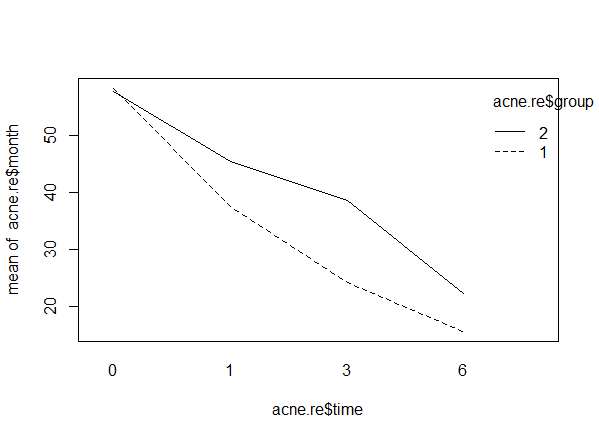
interaction.plot(acne.re$time, acne.re$group, acne.re$month)
> acne <- read.csv("http://commres.net/wiki/_media/r/10_rmanova.csv")
> str(acne)
'data.frame': 14 obs. of 6 variables:
$ group : int 1 1 1 1 1 1 1 2 2 2 ...
$ id : int 1 2 3 4 5 6 7 8 9 10 ...
$ month0: int 60 52 62 58 65 58 53 55 55 60 ...
$ month1: int 41 38 36 34 34 42 38 42 54 55 ...
$ month3: int 25 23 22 21 28 26 25 33 46 46 ...
$ month6: int 16 12 14 13 18 16 21 22 26 23 ...
> acne
group id month0 month1 month3 month6
1 1 1 60 41 25 16
2 1 2 52 38 23 12
3 1 3 62 36 22 14
4 1 4 58 34 21 13
5 1 5 65 34 28 18
6 1 6 58 42 26 16
7 1 7 53 38 25 21
8 2 8 55 42 33 22
9 2 9 55 54 46 26
10 2 10 60 55 46 23
11 2 11 63 45 40 25
12 2 12 52 41 35 22
13 2 13 61 38 32 18
14 2 14 58 43 39 21
>
> # install.packages("reshape")
> library(reshape)
Warning message:
패키지 ‘reshape’는 R 버전 4.1.3에서 작성되었습니다
>
> acne.re <- reshape(acne, direction="long", varying=3:6, sep="")
>
> str(acne.re)
'data.frame': 56 obs. of 4 variables:
$ group: int 1 1 1 1 1 1 1 2 2 2 ...
$ id : int 1 2 3 4 5 6 7 8 9 10 ...
$ time : num 0 0 0 0 0 0 0 0 0 0 ...
$ month: int 60 52 62 58 65 58 53 55 55 60 ...
- attr(*, "reshapeLong")=List of 4
..$ varying:List of 1
.. ..$ month: chr [1:4] "month0" "month1" "month3" "month6"
.. ..- attr(*, "v.names")= chr "month"
.. ..- attr(*, "times")= num [1:4] 0 1 3 6
..$ v.names: chr "month"
..$ idvar : chr "id"
..$ timevar: chr "time"
> acne.re$group <- factor(acne.re$group)
> acne.re$id <- factor(acne.re$id)
> acne.re$time <- factor(acne.re$time)
> str(acne.re)
'data.frame': 56 obs. of 4 variables:
$ group: Factor w/ 2 levels "1","2": 1 1 1 1 1 1 1 2 2 2 ...
$ id : Factor w/ 14 levels "1","2","3","4",..: 1 2 3 4 5 6 7 8 9 10 ...
$ time : Factor w/ 4 levels "0","1","3","6": 1 1 1 1 1 1 1 1 1 1 ...
$ month: int 60 52 62 58 65 58 53 55 55 60 ...
- attr(*, "reshapeLong")=List of 4
..$ varying:List of 1
.. ..$ month: chr [1:4] "month0" "month1" "month3" "month6"
.. ..- attr(*, "v.names")= chr "month"
.. ..- attr(*, "times")= num [1:4] 0 1 3 6
..$ v.names: chr "month"
..$ idvar : chr "id"
..$ timevar: chr "time"
>
> acne.re
group id time month
1.0 1 1 0 60
2.0 1 2 0 52
3.0 1 3 0 62
4.0 1 4 0 58
5.0 1 5 0 65
6.0 1 6 0 58
7.0 1 7 0 53
8.0 2 8 0 55
9.0 2 9 0 55
10.0 2 10 0 60
11.0 2 11 0 63
12.0 2 12 0 52
13.0 2 13 0 61
14.0 2 14 0 58
1.1 1 1 1 41
2.1 1 2 1 38
3.1 1 3 1 36
4.1 1 4 1 34
5.1 1 5 1 34
6.1 1 6 1 42
7.1 1 7 1 38
8.1 2 8 1 42
9.1 2 9 1 54
10.1 2 10 1 55
11.1 2 11 1 45
12.1 2 12 1 41
13.1 2 13 1 38
14.1 2 14 1 43
1.3 1 1 3 25
2.3 1 2 3 23
3.3 1 3 3 22
4.3 1 4 3 21
5.3 1 5 3 28
6.3 1 6 3 26
7.3 1 7 3 25
8.3 2 8 3 33
9.3 2 9 3 46
10.3 2 10 3 46
11.3 2 11 3 40
12.3 2 12 3 35
13.3 2 13 3 32
14.3 2 14 3 39
1.6 1 1 6 16
2.6 1 2 6 12
3.6 1 3 6 14
4.6 1 4 6 13
5.6 1 5 6 18
6.6 1 6 6 16
7.6 1 7 6 21
8.6 2 8 6 22
9.6 2 9 6 26
10.6 2 10 6 23
11.6 2 11 6 25
12.6 2 12 6 22
13.6 2 13 6 18
14.6 2 14 6 21
>
> attach(acne.re)
The following objects are masked from acne.re (pos = 4):
group, id, month, time
> acne.re.anova <- aov(month~group*time+Error(id/time), data=acne.re)
> summary(acne.re.anova)
Error: id
Df Sum Sq Mean Sq F value Pr(>F)
group 1 707.2 707.2 19.71 0.000808 ***
Residuals 12 430.6 35.9
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: id:time
Df Sum Sq Mean Sq F value Pr(>F)
time 3 11366 3789 308.78 < 2e-16 ***
group:time 3 396 132 10.77 3.4e-05 ***
Residuals 36 442 12
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
> interaction.plot(acne.re$time, acne.re$group, acne.re$month)
>
E.g. 3
demo1 <- read.csv("https://stats.idre.ucla.edu/stat/data/demo1.csv")
demo1
str(demo1) ## 모든 변인이 int이므로 (숫자) factor로 바꿔야 한다
## Convert variables to factor
demo1 <- within(demo1, {
group <- factor(group)
time <- factor(time)
id <- factor(id)
}) ## 이제 pulse만 제외하고 모두 factor로 변환된 데이터
str(demo1)
par(cex = .6)
with(demo1, interaction.plot(time, group, pulse,
ylim = c(5, 20), lty= c(1, 12), lwd = 3,
ylab = "mean of pulse", xlab = "time", trace.label = "group"))
demo1.aov <- aov(pulse ~ group * time + Error(id), data = demo1)
summary(demo1.aov)
> summary(demo1.aov)
Error: id
Df Sum Sq Mean Sq F value Pr(>F)
group 1 155.04 155.04 3721 1.3e-09 ***
Residuals 6 0.25 0.04
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
time 2 0.0833 0.04167 1 0.397
group:time 2 0.0833 0.04167 1 0.397
Residuals 12 0.5000 0.04167
demo2
demo2 <- read.csv("https://stats.idre.ucla.edu/stat/data/demo2.csv")
## Convert variables to factor
demo2 <- within(demo2, {
group <- factor(group)
time <- factor(time)
id <- factor(id)
})
demo2
with(demo2, interaction.plot(time, group, pulse,
ylim = c(10, 40), lty = c(1, 12), lwd = 3,
ylab = "mean of pulse", xlab = "time", trace.label = "group"))
demo2.aov <- aov(pulse ~ group * time + Error(id), data = demo2)
summary(demo2.aov)
> demo2 <- read.csv("https://stats.idre.ucla.edu/stat/data/demo2.csv")
> ## Convert variables to factor
> demo2 <- within(demo2, {
+ group <- factor(group)
+ time <- factor(time)
+ id <- factor(id)
+ })
> demo2
id group pulse time
1 1 1 14 1
2 1 1 19 2
3 1 1 29 3
4 2 1 15 1
5 2 1 25 2
6 2 1 26 3
7 3 1 16 1
8 3 1 16 2
9 3 1 31 3
10 4 1 12 1
11 4 1 24 2
12 4 1 32 3
13 5 2 10 1
14 5 2 21 2
15 5 2 24 3
16 6 2 17 1
17 6 2 26 2
18 6 2 35 3
19 7 2 19 1
20 7 2 22 2
21 7 2 32 3
22 8 2 15 1
23 8 2 23 2
24 8 2 34 3
>
> with(demo2, interaction.plot(time, group, pulse,
+ ylim = c(10, 40), lty = c(1, 12), lwd = 3,
+ ylab = "mean of pulse", xlab = "time", trace.label = "group"))
>
> demo2.aov <- aov(pulse ~ group * time + Error(id), data = demo2)
> summary(demo2.aov)
Error: id
Df Sum Sq Mean Sq F value Pr(>F)
group 1 15.04 15.04 0.836 0.396
Residuals 6 107.92 17.99
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
time 2 978.2 489.1 53.684 1.03e-06 ***
group:time 2 1.1 0.5 0.059 0.943
Residuals 12 109.3 9.1
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
demo 3
demo3 <- read.csv("https://stats.idre.ucla.edu/stat/data/demo3.csv")
## Convert variables to factor
demo3 <- within(demo3, {
group <- factor(group)
time <- factor(time)
id <- factor(id)
})
with(demo3, interaction.plot(time, group, pulse,
ylim = c(10, 60), lty = c(1, 12), lwd = 3,
ylab = "mean of pulse", xlab = "time", trace.label = "group"))
demo3.aov <- aov(pulse ~ group * time + Error(id), data = demo3)
summary(demo3.aov)
> demo3 <- read.csv("https://stats.idre.ucla.edu/stat/data/demo3.csv")
> ## Convert variables to factor
> demo3 <- within(demo3, {
+ group <- factor(group)
+ time <- factor(time)
+ id <- factor(id)
+ })
>
> with(demo3, interaction.plot(time, group, pulse,
+ ylim = c(10, 60), lty = c(1, 12), lwd = 3,
+ ylab = "mean of pulse", xlab = "time", trace.label = "group"))
>
> demo3.aov <- aov(pulse ~ group * time + Error(id), data = demo3)
> summary(demo3.aov)
Error: id
Df Sum Sq Mean Sq F value Pr(>F)
group 1 2035.0 2035.0 343.1 1.6e-06 ***
Residuals 6 35.6 5.9
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
time 2 2830.3 1415.2 553.8 1.52e-12 ***
group:time 2 200.3 100.2 39.2 5.47e-06 ***
Residuals 12 30.7 2.6
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
>
r/twoway_repeated_measure_anova.txt · Last modified: by hkimscil